Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. [Nucleic Acids Res 48: e74(https://doi.org/10.1093/nar/gkaa426)).
See the 2020 paper titled "DSSR-enabled innovative schematics of 3D nucleic acid structures with PyMOL" in Nucleic Acids Research and the corresponding Supplemental PDF for details. Many thanks to Drs. Wilma Olson and Cathy Lawson for their help in the preparation of the illustrations.
Details on how to reproduce the cover images are available on the 3DNA Forum.

Structure of a group II intron ribonucleoprotein in the pre-ligation state (PDB id: 8T2R; Xu L, Liu T, Chung K, Pyle AM. 2023. Structural insights into intron catalysis and dynamics during splicing. Nature 624: 682–688). The pre-ligation complex of the Agathobacter rectalis group II intron reverse transcriptase/maturase with intron and 5′-exon RNAs makes it possible to construct a picture of the splicing active site. The intron is depicted by a green ribbon, with bases and Watson-Crick base pairs represented as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; the 5′-exon is shown by white spheres and the protein by a gold ribbon. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Complex of terminal uridylyltransferase 7 (TUT7) with pre-miRNA and Lin28A (PDB id: 8OPT; Yi G, Ye M, Carrique L, El-Sagheer A, Brown T, Norbury CJ, Zhang P, Gilbert RJ. 2024. Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs. Nat Struct Mol Biol 31: 1426–1438). The RNA-binding pluripotency factor LIN28A invades and melts the RNA and affects the mechanism of action of the TUT7 enzyme. The RNA backbone is depicted by a red ribbon, with bases and Watson-Crick base pairs represented as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; TUT7 is represented by a gold ribbon and LIN28A by a white ribbon. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Cryo-EM structure of the pre-B complex (PDB id: 8QP8; Zhang Z, Kumar V, Dybkov O, Will CL, Zhong J, Ludwig SE, Urlaub H, Kastner B, Stark H, Lührmann R. 2024. Structural insights into the cross-exon to cross-intron spliceosome switch. Nature 630: 1012–1019). The pre-B complex is thought to be critical in the regulation of splicing reactions. Its structure suggests how the cross-exon and cross-intron spliceosome assembly pathways converge. The U4, U5, and U6 snRNA backbones are depicted respectively by blue, green, and red ribbons, with bases and Watson-Crick base pairs shown as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; the proteins are represented by gold ribbons. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Structure of the Hendra henipavirus (HeV) nucleoprotein (N) protein-RNA double-ring assembly (PDB id: 8C4H; Passchier TC, White JB, Maskell DP, Byrne MJ, Ranson NA, Edwards TA, Barr JN. 2024. The cryoEM structure of the Hendra henipavirus nucleoprotein reveals insights into paramyxoviral nucleocapsid architectures. Sci Rep 14: 14099). The HeV N protein adopts a bi-lobed fold, where the N- and C-terminal globular domains are bisected by an RNA binding cleft. Neighboring N proteins assemble laterally and completely encapsidate the viral genomic and antigenomic RNAs. The two RNAs are depicted by green and red ribbons. The U bases of the poly(U) model are shown as cyan blocks. Proteins are represented as semitransparent gold ribbons. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Structure of the helicase and C-terminal domains of Dicer-related helicase-1 (DRH-1) bound to dsRNA (PDB id: 8T5S; Consalvo CD, Aderounmu AM, Donelick HM, Aruscavage PJ, Eckert DM, Shen PS, Bass BL. 2024. Caenorhabditis elegans Dicer acts with the RIG-I-like helicase DRH-1 and RDE-4 to cleave dsRNA. eLife 13: RP93979. Cryo-EM structures of Dicer-1 in complex with DRH-1, RNAi deficient-4 (RDE-4), and dsRNA provide mechanistic insights into how these three proteins cooperate in antiviral defense. The dsRNA backbone is depicted by green and red ribbons. The U-A pairs of the poly(A)·poly(U) model are shown as long rectangular cyan blocks, with minor-groove edges colored white. The ADP ligand is represented by a red block and the protein by a gold ribbon. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).
Moreover, the following 30 [12(2021) + 12(2022) + 6(2023)] cover images of the RNA Journal were generated by the NAKB (nakb.org).
Cover image provided by the Nucleic Acid Database (NDB)/Nucleic Acid Knowledgebase (NAKB; nakb.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).
In early 2015, Thomas Holder (the PyMOL Principal Developer at Schrodinger) and I agreed to work together on connecting DSSR to PyMOL. Moreover, we called for the community’s involvement in writing a DSSR plugin for PyMOL and received a few enthusiastic replies. Over the past few months, many significant progresses have been made in DSSR, including an article titled DSSR: an integrated software tool for dissecting the spatial structure of RNA published in Nucleic Acids Research (NAR) and a more streamlined DSSR-Jmol integration based on the --json output.
From the very beginning, Thomas and I had envisioned that the DSSR-PyMOL integration would include two components: one is to bring DSSR-derived RNA/DNA structural features into PyMOL (similar to the DSSR-Jmol interface, funcationality-wise), and the other is to render DSSR’s simple yet informative base-rectangular representations with PyMOL. While the ‘analysis’ component is a work in progress, the ‘visualization’ part is ready for the community to take advantage of.
Thomas has written a Python script named dssr_block.py. When the script is run in PyMOL, it adds the “dssr_block” command. The dssr_block.py script is less than 100 lines including documentation, with the real code taking no more than half of the total line number. The detailed documentation section (with two examples), when condensed, is as follows:
DESCRIPTION
Create a nucleid acid cartoon with DSSR
USAGE
dssr_block [selection [, state [, block_file [, block_depth [, name [, exe]]]]]]
ARGUMENTS
selection = str: atom selection {default: all}
state = int: object state (0 for all states) {default: -1, current state}
block_file = face|edge|wc|equal|minor|gray {default: face}
block_depth = float: thickness of rectangular blocks {default: 0.5}
name = str: name of new CGO object {default: dssr_block##}
exe = str: path to "x3dna-dssr" executable {default: x3dna-dssr}
EXAMPLE
fetch 1ehz, async=0
as cartoon
dssr_block
set cartoon_ladder_radius, 0.1
set cartoon_ladder_color, gray
set cartoon_nucleic_acid_mode, 1
# multi-state
fetch 2n2d, async=0
dssr_block 2n2d, 0
set all_states
Download the dssr_block.py script into a folder (directory) of your choice. Within PyMOL command window, type:
run dssr_block.py # to make the 'dssr_block' command avaible
help dssr_block # to get the help message, with contents shown above
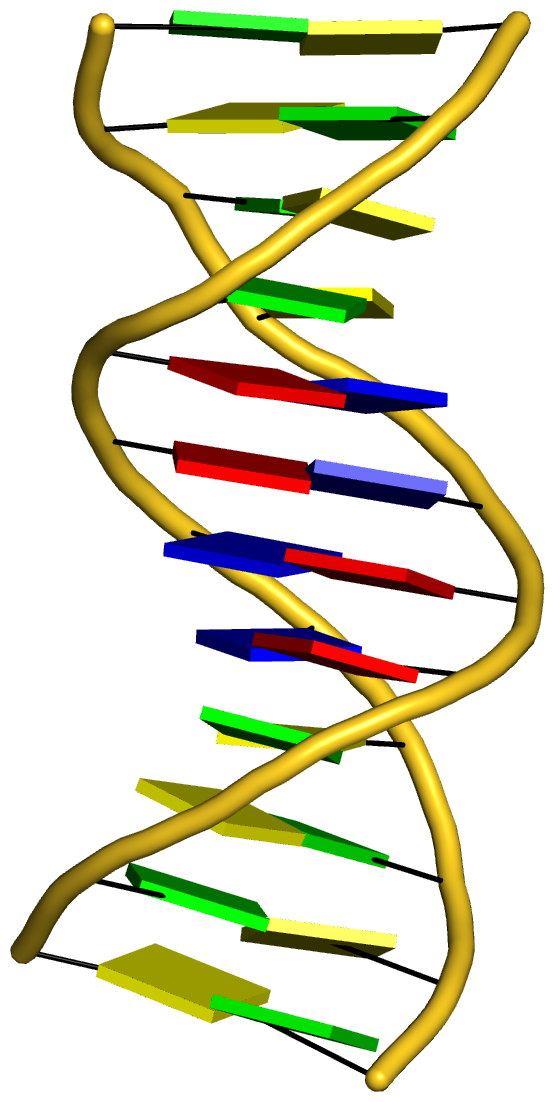
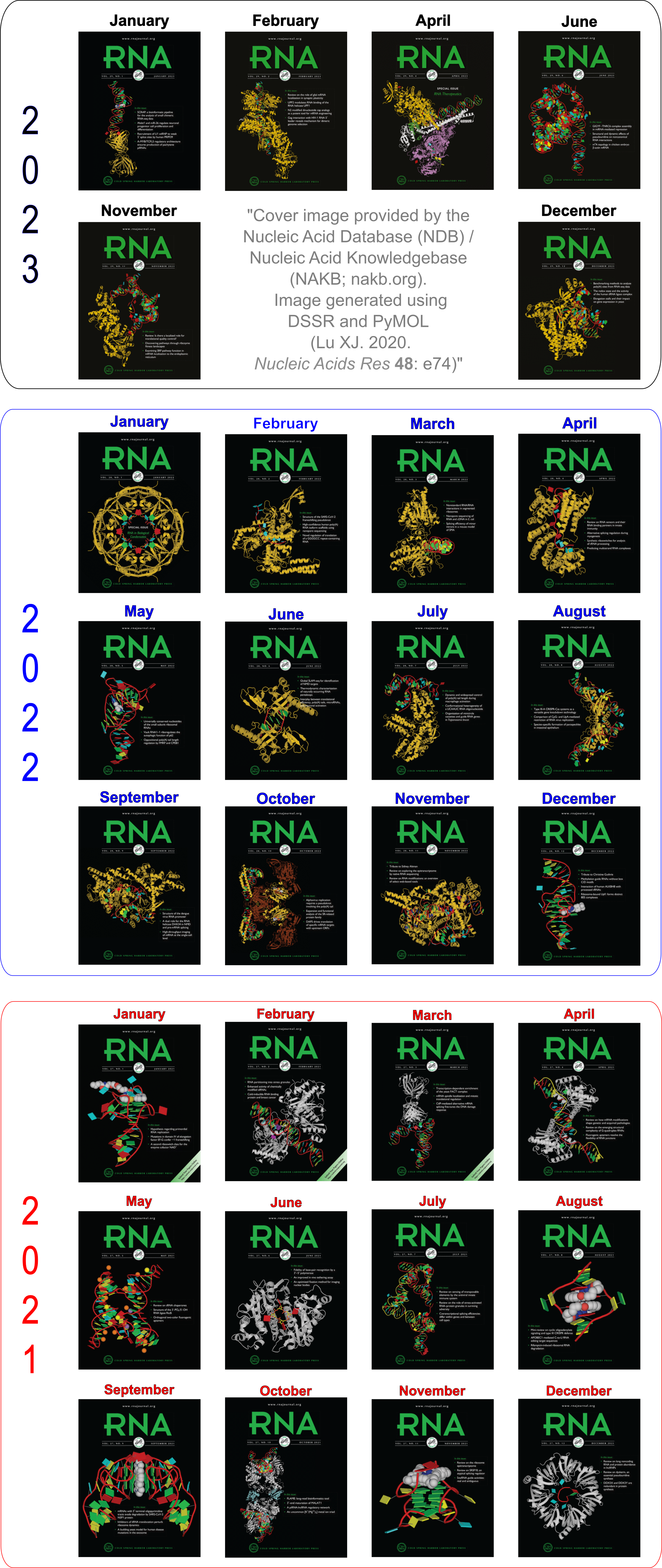
The resultant cartoon-block image for running the documented commands (except for the additional orient command for best view) for case 1ehz is shown in Fig. 1 below.
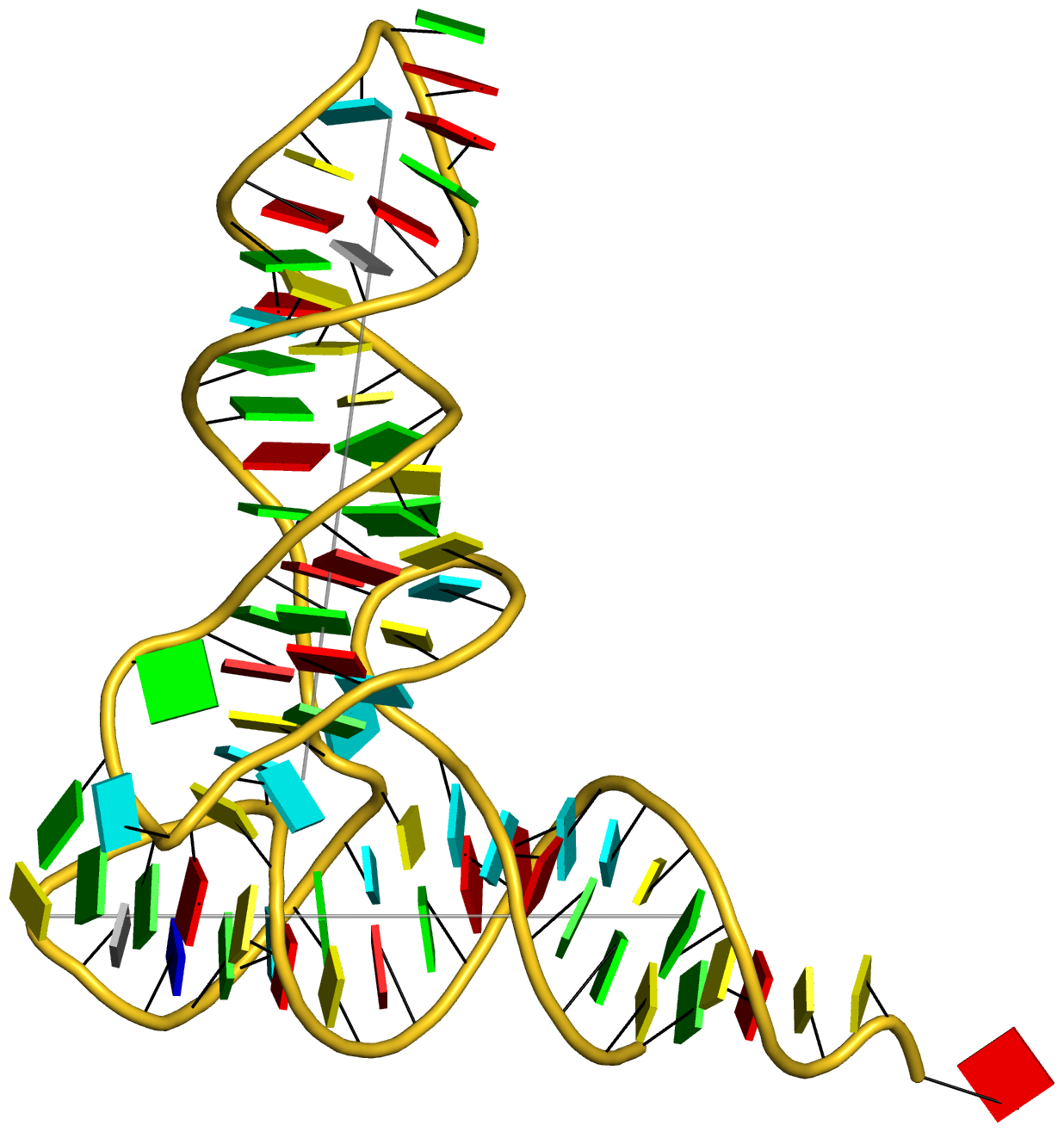
Fig. 1: Cartoon-block image generated by dssr_block.py for PDB entry 1ehz (yeast phenylalanine tRNA)
For the NMR ensemble 2n2d, the corresponding image (after running orient) is illustrated in Fig. 2 as follows:
Fig. 2: Cartoon-block image generated by dssr_block.py for PDB entry 2n2d (an NMR ensemble).
In addition to the default settings, DSSR offers quite a few variations for the size and coloring of rectangular blocks, as demonstrated in Fig.3. The main settings are through the block_file option in PyMOL (note the underscore), corresponding to DSSR --block-file (or --block_file). The corresponding PyMOL commands are also listed for your reference. You can easily play around with the various styles interactively in PyMOL by toggling objects (dssr_block##) on or off. Enjoy!
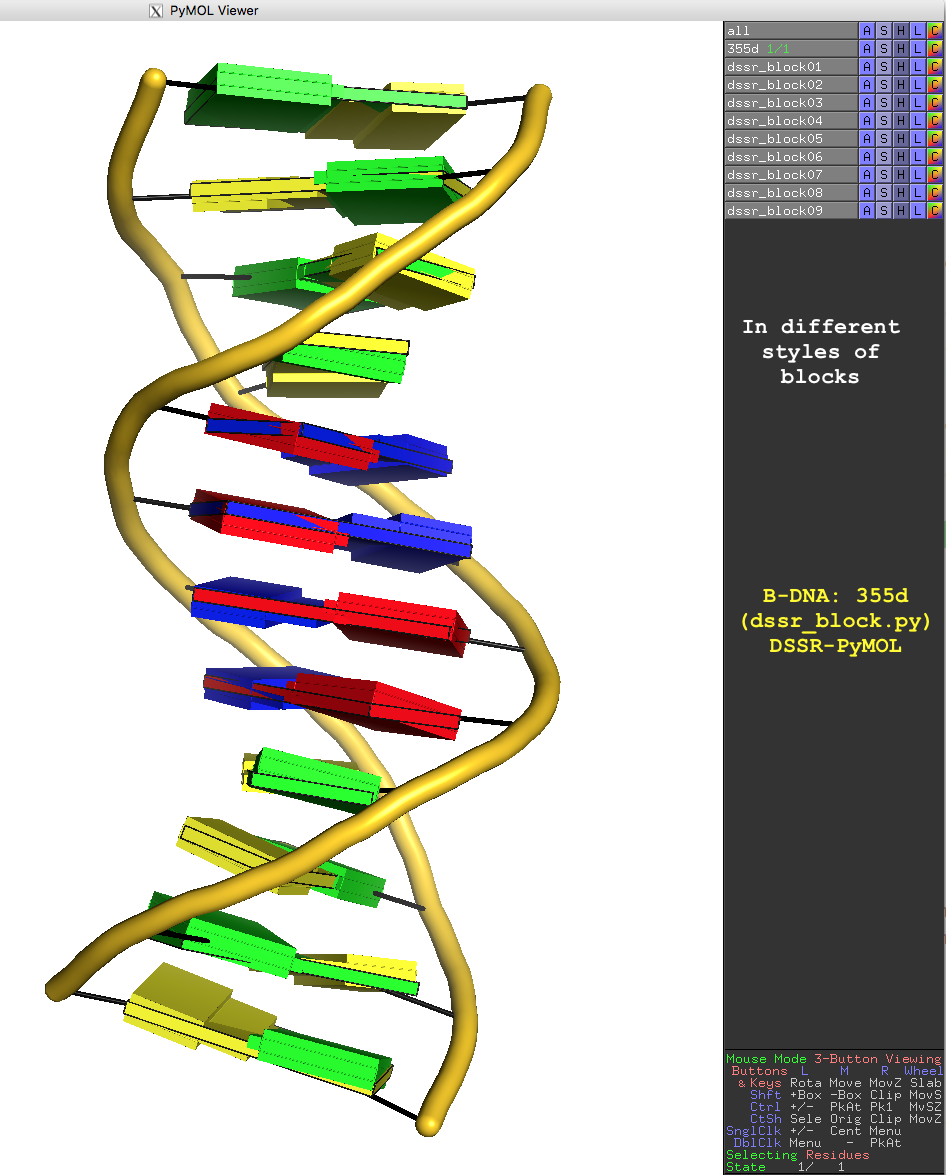
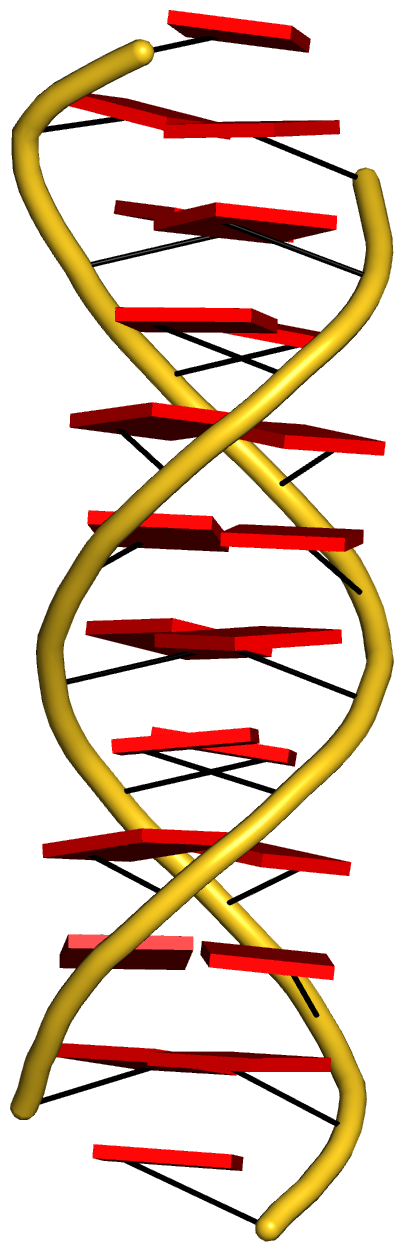
Fig. 3: Cartoon-block image generated by dssr_block.py for PDB entry 355d (the Dickerson B-DNA dodecamer).
Fig. 3 is created with the following PyMOL commands:
reinitialize
fetch 355d, async=0
bg_color white
as cartoon
orient
turn z, -90
turn y, 180
set cartoon_ladder_mode, 1
set cartoon_ladder_radius, 0.1
set cartoon_ladder_color, black
set cartoon_tube_radius, 0.5
set cartoon_nucleic_acid_mode, 1
set cartoon_color, gold
dssr_block 355d # default base blocks in solid color
dssr_block block_file=edge # rectangular blocks in wireframe (black)
dssr_block block_file=face+edge # solid color with outline
dssr_block block_file=equal # bases blocks in equal size
dssr_block block_file=minor # with minor-groove colord black
dssr_block block_file=wc # Watson-Crick base pairs in long bp blocks
dssr_block block_file=wc-minor # Watson-Crick pairs + minor-groove edge
dssr_block block_file=gray # rectangular blocks all in gray
dssr_block block_depth=1.8 # with increased thickness
Notes
- The
dssr_block.py script described here is the original version Thomas communicated to me. Current version of this script and related topics can be found in the Dssr block PyMOLWiki page.
- For this script to work, DSSR needs to be installed and
x3dna-dssr in the PATH.
- In PyMOL,
set cartoon_nucleic_acid_mode, 1 employs C3′ instead of the default P (‘mode 0’) for the smooth backbone trace. Since 5′ terminal phosphate groups are normally not available from X-ray crystal structures (e.g., 355d), ‘mode 1’ is used to avoid orphan base blocks from the backbone trace.

With the foundation laid by the previous two posts on Fitting of base reference frame and Automatic identification of nucleotides, we can now get into the details on how the ‘simple’ base-pair (bp) parameters are derived. To make the point clear, I am using two concrete examples from the yeast phenylalanine tRNA (PDB id: 1ehz): the first pair is 2MG10+G45, of type M+N (shortened to g+G) in 3DNA/DSSR; and the second example is a Watson-Crick pair U6–A67, of type M–N (shortened to U–A).
Pair 2MG10+G45 (g+G, of type M+N, see Fig. 1)
Base reference frames
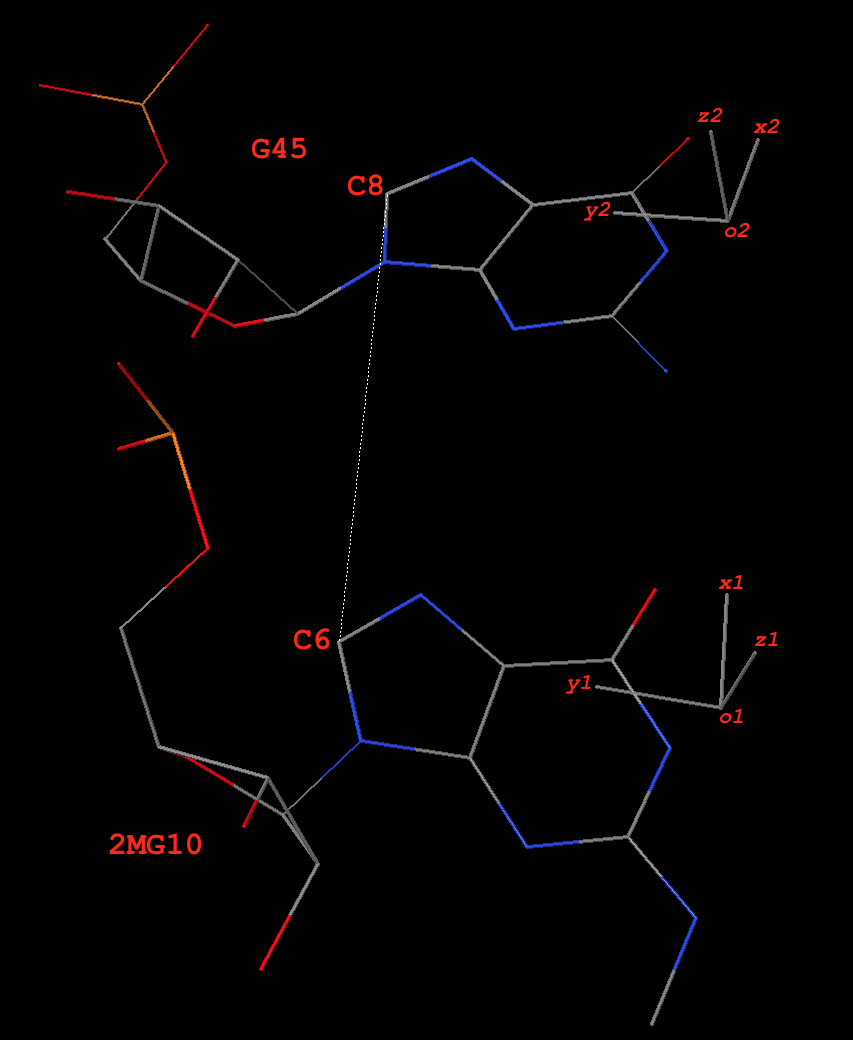
Fig. 1: Base pair 2MG10+G45 (g+G) of type M+N in yeast phenylalanine tRNA 1ehz
In the original coordinate system (as in 1ehz.pdb downloaded from the RCSB PDB), the base-reference frames for 2MG10 and G45 are:
# base reference frame of 2MG10
{
"rsmd": 0.018218,
"origin": [65.696016, 45.134944, 18.125044], # o1
"x_axis": [0.690346, 0.713907, -0.117302], # x1
"y_axis": [-0.706849, 0.700116, 0.101003], # y1
"z_axis": [0.154232, 0.013188, 0.987947] # z1
}
# base reference frame of G45
{
"rsmd": 0.025865,
"origin": [70.584399, 50.526567, 17.229626], # o2
"x_axis": [0.818521, 0.49914, -0.284399], # x2
"y_axis": [-0.574112, 0.728382, -0.373973], # y2
"z_axis": [0.020486, 0.469381, 0.882758] # z2
}
The base-pair reference frame
Since dot(z1, z2) = 0.88 (positive), this pair is of type M+N in 3DNA/DSSR. The ‘mean’ z-axis of the pair is the average of z1 and z2, which is z = [0.090069, 0.248769, 0.964366] (normalized). This is the z-axis of the bp frame, as in 3DNA/DSSR.
The ‘long’ axis employs RC8 (purines) and YC6 (pyrimidines) base atoms. Here 2MG10 and G45 are all purines, so the following two C8 atoms are used:
# C8 atoms of 2MG10 and G45 in 1ehz
HETATM 208 C8 2MG A 10 62.199 48.621 18.635 1.00 40.38 C
ATOM 987 C8 G A 45 67.772 54.149 15.386 1.00 40.45 C
The vector from C8 of G45 to C8 of 2MG10 is:
y0 = [62.199 48.621 18.635] - [67.772 54.149 15.386]
= [-5.573 -5.528 3.249]
Normally, y0 and z-axis are not orthogonal. Here they have an angle of ~81º. The orthogonal component of y0 with reference to the z-axis, when normalized, is the y-axis:
y = [-0.676751, -0.695120, 0.242520]
The x-axis is defined by the right-handed rule:
x = [-0.730682, 0.674479, -0.105746]
Overall, the orthonormal x-, y- and z-axes of the pair defined thus far are:
x = [-0.730682, 0.674479, -0.105746]
y = [-0.676751, -0.695120, 0.242520]
z = [0.090069, 0.248769, 0.964366]
Derivation of the six ‘simple’ base-pair parameters (Fig. 2)
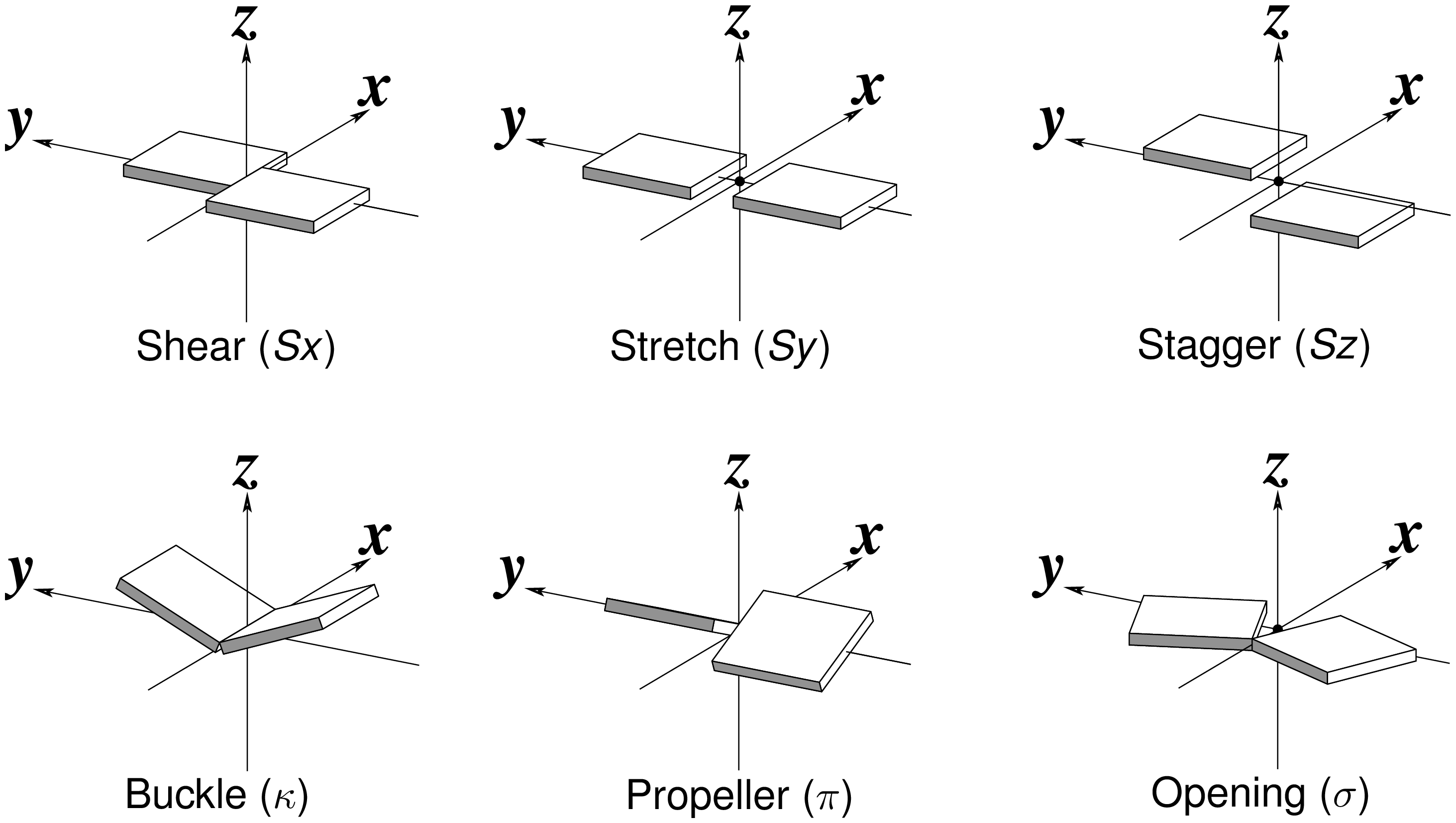
Fig. 2: Schematic diagram of six rigid-body base-pair parameters
Propeller is the ‘torsion’ angle of z2 to z1 with reference to the y-axis, and is calculated using the method detailed in the blog post How to calculate torsion angle?. Here Propeller is: -24.24º. Similarly, Buckle is defined as the ‘torsion’ angle of z2 to z1 with reference to the x-axis, and is -14.81º. Opening is defined as the angle from y2 to y1 with reference to the z-axis, and is: 13.32º.
The corresponding translational parameters are simply projects of the o2 to o1 vector onto the x-, y- and z-axis, respectively. Here, they have values:
d = o1 - o2 = [-4.888383, -5.391623, 0.895418]
Shear = dot(d, x) = -0.16
Stretch = dot(d, y) = 7.27
Stagger = dot(d, z) = -0.92
‘Corrections’ of Buckle and Propeller
Base-pair non-planarity is due to the following three parameters: Buckle, Propeller, and Stagger. In particular, Buckle and Propeller cause the two bases to be non-parallel, the most noticeable characteristic of a pair. These two angular parameters are well-documented in literature, even among the canonical Watson-Crick base pairs. In 3DNA/DSSR, the angle between the two base normal vectors (in range [0, 90º]) is related to Buckle and Propeller with the formula:
interBase-angle = sqrt(Buckle^2+Propeller^2)
For the 2MG10+G45 pair, the angle between z1 and z2 is 28.18º, and sqrt(Buckle^2+Propeller^2) = 28.405º. So the following ‘corrections’ are made:
Buckle = -14.81 * 28.18 / 28.405 = -14.69
Propeller = -24.24 * 28.18 / 28.405 = -24.05
Overall, the ‘corrections’ have only small influence on the numerical values of the reported Buckle and Propeller parameters. It is ‘sensible’ that the ‘simple’ parameters have the property interBase-angle = sqrt(Buckle^2+Propeller^2), just as the original 3DNA/DSSR bp parameters.
Now, the six ‘simple’ bp parameters for 2MG10+G45, reported in 3DNA analyze program as of v2.3-2016jan01 are:
Simple base-pair parameters based on YC6-RC8 vectors
bp Shear Stretch Stagger Buckle Propeller Opening angle
* 1 g+G -0.16 7.27 -0.92 -14.69 -24.05 13.32 28.2
The corresponding local bp parameters as originally reported by 3DNA/DSSR are as follows. Note the significant differences in Shear vs. Stretch, and Buckle vs. Propeller in the two sets of bp parameters. On the other hand, Stagger is identical and Opening should be quite close, by definition. Due to the similarity in Stagger and Opening, DSSR only reports four ‘simple’ parameters (i.e., Shear, Stretch, Buckle, and Propeller).
Local base-pair parameters
bp Shear Stretch Stagger Buckle Propeller Opening
1 g+G -7.21 -0.97 -0.92 25.58 -11.83 13.07
Base-pair U6–A67 (Watson-Crick U–A, of type M–N, see Fig. 3)
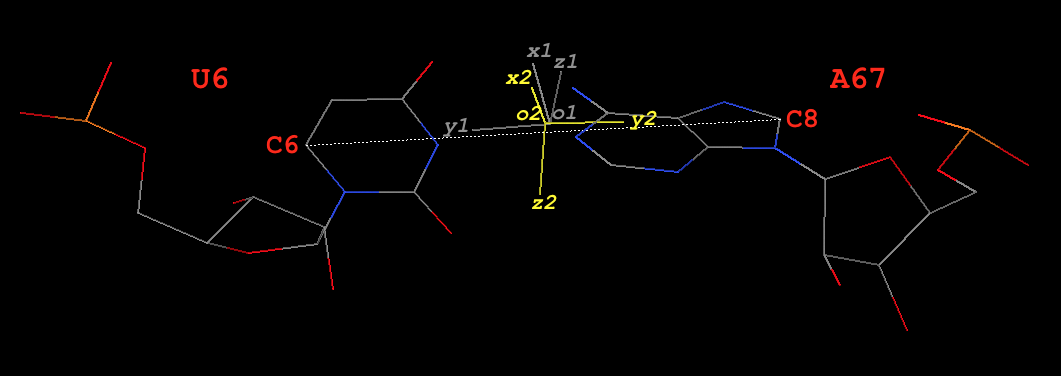
Fig. 3: Base pair U6–A67 (U–A) of type M–N in yeast phenylalanine tRNA 1ehz
Base reference frames
In the original coordinate system (as in 1ehz.pdb downloaded from the RCSB PDB), the base-reference frames for U6 and A67 are:
# base reference frame of U6 (white in Fig. 3)
{
"rsmd": 0.010835,
"origin": [60.441988, 48.83479, 41.242523], # o1
"x_axis": [0.28491, 0.503019, 0.815965], # x1
"y_axis": [0.887155, -0.460753, -0.025726], # y1
"z_axis": [0.363018, 0.731217, -0.577529] # z1
}
# base reference frame of A67 (colored yellow in Fig. 3)
{
"rsmd": 0.01992,
"origin": [60.578326, 48.823104, 41.154211], # o2
"x_axis": [0.034097, 0.205538, 0.978055], # x2
"y_axis": [-0.90687, 0.417653, -0.056155], # y2
"z_axis": [-0.420029, -0.885054, 0.200637] # z2
}
The base-pair reference frame
Since dot(z1, z2) = -0.92 (negative), this pair is of type M–N in 3DNA/DSSR. The y- and z-axis are thus reversed (corresponding to a 180º rotation around the x-axis) to align z2 with z1.
# base reference frame of A67, with y- and z-axes reversed
{
"origin": [60.578326, 48.823104, 41.154211], # o2
"x_axis": [0.034097, 0.205538, 0.978055], # x2
"y_axis": [0.90687, -0.417653, 0.056155], # y2 -- reversed
"z_axis": [0.420029, 0.885054, -0.200637] # z2 -- reversed
}
Thereafter, the procedure is similar to the one for the M+N type above. Note here U6 is a pyrimidine, so its C6 atom is used. The final results are:
# C6 atom of U6 and C8 atom A67 in 1ehz
ATOM 132 C6 U A 6 64.926 46.497 41.084 1.00 35.72 C
ATOM 1457 C8 A A 67 56.129 50.866 40.893 1.00 40.04 C
#---------
y0 = [64.926 46.497 41.084] - [56.129 50.866 40.893]
= [8.797 -4.369 0.191]
x = [0.160777, 0.363836, 0.917482]
y = [0.902274, -0.430972, 0.012793]
z = [0.400064, 0.825764, -0.397570]
The six ‘simple’ and original base-pair parameters
Simple base-pair parameters based on YC6-RC8 vectors
bp Shear Stretch Stagger Buckle Propeller Opening angle
1 U-A 0.06 -0.13 -0.08 -0.59 -23.71 5.39 23.7
# ------------
Local base-pair parameters
bp Shear Stretch Stagger Buckle Propeller Opening
1 U-A 0.06 -0.13 -0.08 -0.63 -23.71 5.50
As can be seen, for Watson-Crick pairs, the ‘simple’ and the original bp parameters are very similar.
Special notes on the ‘simple’ base-pair parameters

- For the most common Watson-Crick pairs, the newly introduced ‘simple’ bp parameters match those of the original 3DNA/DSSR parameters very well (as shown by the U6–A67 pair). For non-canonical pairs, significant differences in Shear, Stretch, Buckle and Propeller are expected (as illustrated by the 2MG10+G45 pair). The differences come from the divergent definitions of the bp reference frame, which is distinct for each type of non-canonical pairs.
- Only the original 3DNA/DSSR six bp parameters can be used for exact reconstruction (with the 3DNA
rebuild program) of the corresponding bp geometry. The ‘simple’ bp parameters are for description only, and they could be more intuitive than the original 3DNA/DSSR counterparts. They complement, buy by no means replace, the classic “local” bp parameters. The term ‘simple’ is used to distinguish the new from the original closely related, yet quite different bp parameters.
- As details for the 2MG10+G45 pair, several ad hoc decisions are made in deriving the ‘simple’ bp parameters. For example, instead of using RC8–YC6 to define the y-axis, one can also use RN9–YN1 (as did by Richardson). Each such choice will lead (slightly) different numerical values, depending on the type of the non-canonical pairs. In some cases, Buckle and Propeller could differ by several degrees. Since RC8 and YC6 atoms lie near the ‘center’ of purines and pyrimidines, they are used to define the y-axis (by default). DSSR has provisions of selecting RN9–YN1, as well as a couple of other choices, for the definition of the y-axis.
- When the M+N pair is counted as N+M, Shear, Stretch, Buckle, and Propeller remain the same, but Stagger and Opening reverse their signs. For example, here are the results of 2MG10+G45 vs. G45+2MG10:
# 2MG10+G45
Simple base-pair parameters based on YC6-RC8 vectors
bp Shear Stretch Stagger Buckle Propeller Opening angle
* 1 G+g -0.16 7.27 0.92 -14.69 -24.05 -13.32 28.2
# Reverse the order: treated as G45+2MG10
Simple base-pair parameters based on YC6-RC8 vectors
bp Shear Stretch Stagger Buckle Propeller Opening angle
* 1 g+G -0.16 7.27 -0.92 -14.69 -24.05 13.32 28.2
- When the M–N pair is counted as N–M, Stretch, Stagger, Propeller, and Opening remain the same, but Shear and Buckle reverse their signs. For example, here are the results of U6–A67 vs. A67–U6:
# U6–A67
Simple base-pair parameters based on YC6-RC8 vectors
bp Shear Stretch Stagger Buckle Propeller Opening angle
1 U-A 0.06 -0.13 -0.08 -0.59 -23.71 5.39 23.7
# Reverse the order: treated as A67–U6
Simple base-pair parameters based on YC6-RC8 vectors
bp Shear Stretch Stagger Buckle Propeller Opening angle
1 A-U -0.06 -0.13 -0.08 0.59 -23.71 5.39 23.7
Related posts

Once a nucleotide (nt) is identified, and matched to A (C, G, T, U) for the standard case or a (c, g, t, u) for a modified one, 3DNA/DSSR performs a least-squares fitting procedure to locate the base reference frame in three-dimensional space. The basic idea is very simple and widely applicable. The algorithm constitutes one of the key components of 3DNA/DSSR. As always, the details can be most effectively illustrated with a worked example. Using G1 in the yeast phenylalanine tRNA (PDB id: 1ehz) as an example, the atomic coordinates of its nine base-ring atoms are:
# G1, nine base-ring atoms for ls-fitting
ATOM 14 N9 G A 1 51.628 45.992 53.798 1.00 93.67 N
ATOM 15 C8 G A 1 51.064 46.007 52.547 1.00 92.60 C
ATOM 16 N7 G A 1 51.379 44.966 51.831 1.00 91.19 N
ATOM 17 C5 G A 1 52.197 44.218 52.658 1.00 91.47 C
ATOM 18 C6 G A 1 52.848 42.992 52.425 1.00 90.68 C
ATOM 20 N1 G A 1 53.588 42.588 53.534 1.00 90.71 N
ATOM 21 C2 G A 1 53.685 43.282 54.716 1.00 91.21 C
ATOM 23 N3 G A 1 53.077 44.429 54.946 1.00 91.92 N
ATOM 24 C4 G A 1 52.356 44.836 53.879 1.00 92.62 C
The corresponding nine base-ring atoms of G in its standard base reference frame are listed below. See Table 1 of the report A Standard Reference Frame for the Description of Nucleic Acid Base-pair Geometry, and file Atomic_G.pdb distributed with 3DNA ($X3DNA/config/Atomic_G.pdb). In DSSR, the content has been integrated into the source code to make the program self-contained.
# G in standard base reference frame
ATOM 2 N9 G A 1 -1.289 4.551 0.000
ATOM 3 C8 G A 1 0.023 4.962 0.000
ATOM 4 N7 G A 1 0.870 3.969 0.000
ATOM 5 C5 G A 1 0.071 2.833 0.000
ATOM 6 C6 G A 1 0.424 1.460 0.000
ATOM 8 N1 G A 1 -0.700 0.641 0.000
ATOM 9 C2 G A 1 -1.999 1.087 0.000
ATOM 11 N3 G A 1 -2.342 2.364 0.001
ATOM 12 C4 G A 1 -1.265 3.177 0.000
A least-squares fitting of the standard onto the experimental set of base-ring atoms defines the base reference frame (Fig. 1). The information is available via the following commands:
# find_pair -s 1ehz.pdb # in file 'ref_frames.dat'
... 1 G # A:...1_:[..G]G
53.7571 41.8678 52.9303 # origin
-0.2589 -0.2496 -0.9331 # x-axis
-0.5430 0.8365 -0.0731 # y-axis
0.7988 0.4878 -0.3521 # z-axis
# --------
# x3dna-dssr -i=1ehz.pdb --json | jq .nts[0].frame
{
rsmd: 0.008,
origin: [53.757, 41.868, 52.93],
x_axis: [-0.259, -0.25, -0.933],
y_axis: [-0.543, 0.837, -0.073],
z_axis: [0.799, 0.488, -0.352]
}
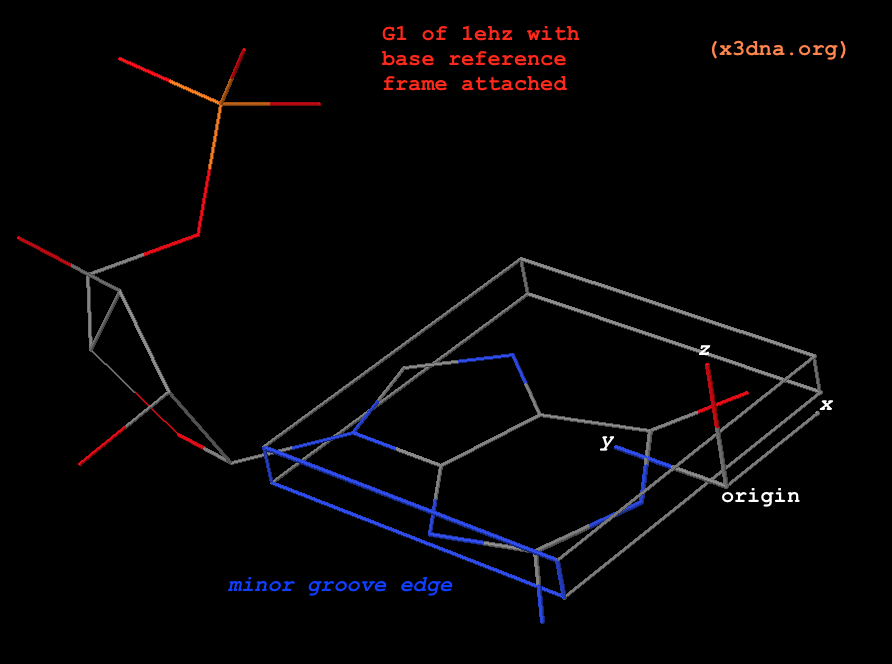
Fig. 1: G1 in tRNA 1ehz, with base reference frame attached
Please note the following subtle points:
- The standard base (
Atomic_G.pdb) is already set in its reference frame: the _z_-coordinates are virtually zeros, _y_-coordinates are positive, the atoms along the minor-groove edge have negative _x_-coordinates, as can be visualized clearly from the attached coordinate frame. In 3DNA, the five standard standard bases are in stored in files Atomic_[ACGTU].pdb, and the corresponding modified ones are in Atomic_[acgtu].pdb. For simplicity, Atomic_A.pdb and Atomic_a.pdb are the same by default, as are the other four cases.
- The translation and rotation of the least-squares fitting process define the experimental base reference frame (for G1 in the above example), and its three axes are orthonormal by definition.
- By design, the base rings of
Atomic_A.pdb and Atomic_G.pdb match each other closely (see below), as are the pyrimidines bases. The least-square fitted root-mean-square deviation (rmsd) of the nine base-ring atoms between standard A and G is only 0.04 Å. Fitting the standard A (instead of G) onto G1 of 1ehz leads to a base reference frame that is essentially indistinguishable from the one above (see below). This feature shows that any ambiguity in assigning modified purines to A or G, or pyrimidines to C, T, or U causes no notable differences in 3DNA/DSSR results.
Comparison of base-ring atomic coordinates in standard G and A
Atomic_G.pdb Atomic_A.pdb
N9 G -1.289 4.551 0.000 | N9 A -1.291 4.498 0.000
C8 G 0.023 4.962 0.000 | C8 A 0.024 4.897 0.000
N7 G 0.870 3.969 0.000 | N7 A 0.877 3.902 0.000
C5 G 0.071 2.833 0.000 | C5 A 0.071 2.771 0.000
C6 G 0.424 1.460 0.000 | C6 A 0.369 1.398 0.000
N1 G -0.700 0.641 0.000 | N1 A -0.668 0.532 0.000
C2 G -1.999 1.087 0.000 | C2 A -1.912 1.023 0.000
N3 G -2.342 2.364 0.001 | N3 A -2.320 2.290 0.000
C4 G -1.265 3.177 0.000 | C4 A -1.267 3.124 0.000
Comparison of G1 (1ehz) base reference frame derived using standard G or A
Atomic_G.pdb | Atomic_A.pdb
53.7571 41.8678 52.9303 # origin | 53.7286 41.9276 52.9482 # origin
-0.2589 -0.2496 -0.9331 # x-axis | -0.2562 -0.2540 -0.9327 # x-axis
-0.5430 0.8365 -0.0731 # y-axis | -0.5444 0.8352 -0.0780 # y-axis
0.7988 0.4878 -0.3521 # z-axis | 0.7988 0.4878 -0.3522 # z-axis
Related topics:

Any analysis of nucleic acid structures start with the identification of nucleotides (nts), the basic building unit. As per the PDB convention, each nt (like any other ligands) is specified by a three-letter identifier. For example, the four standard RNA nts are ..A, ..C, ..G, and ..U, respectively. The four corresponding standard DNA nts are .DA, .DC, .DG, and .DT, respectively. Note that here, for visualization purpose, each space is represented by a dot (.). In practice, the following codes for the five standard DNA/RNA nts — ADE, CYT, GUA, THY, and URA — are also commonly encountered, among other variants.
On top of the standard nts, there are numerous modified ones, each assigned a unique three-letter code. In the classic yeast phenylalanine tRNA (PDB id: 1ehz), 14 out of the 76 nts are modified, as shown in Fig. 1 below.
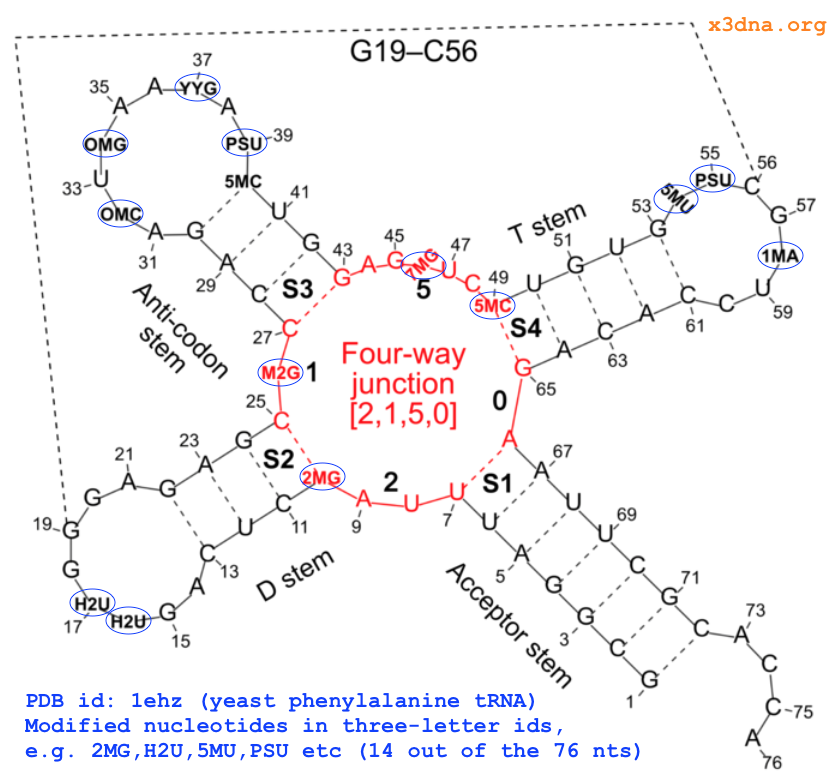
Fig. 1: Modified nucleotides in yeast phenylalanine tRNA 1ehz
It is challenging to maintain a comprehensive and updated list of ever-inceasing nts encountered in the PDB and molecular dynamics (MD) simulation packages (e.g., AMBER, GROMACS, and CHARMM). Thus, as of today, some well-known DNA/RNA structural bioinformatics tools can handle only standard nts or a limited list of modified ones.
From early on in the development of 3DNA, I observed that all recognized nts have a core six-membered ring, with atoms named N1,C2,N3,C4,C5,C6 consecutively (see Fig. 2 below). Purines have three additional atoms, named N7,C8,N9. So it is feasible to automatically identify nts, and classify them as pyrimidines and purines, based on the common core skeleton shared by all of them. Moreover, the ‘skeleton’ is not effected by any possible tautomeric or protonation state.
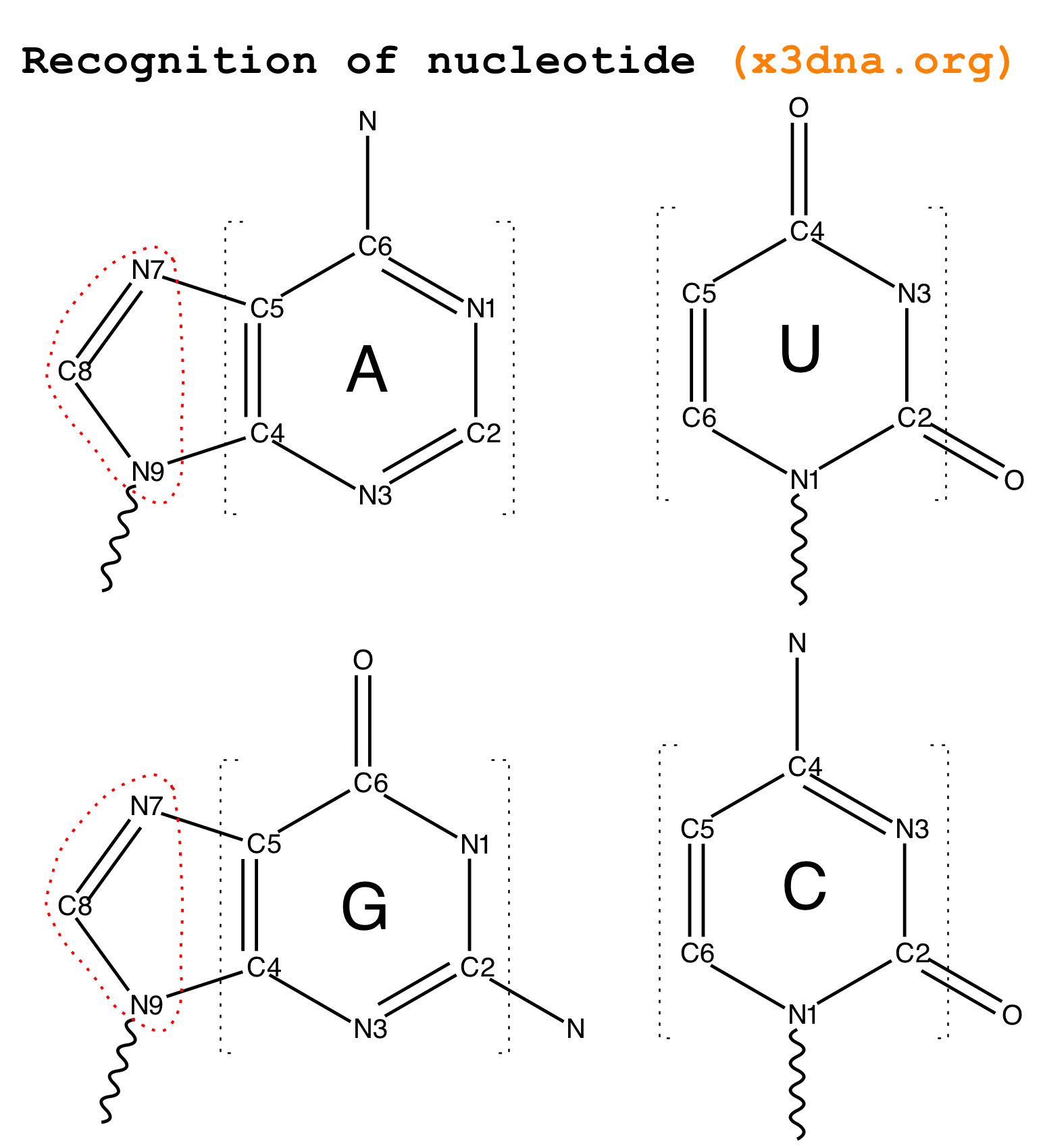
Fig. 2: Identification of nts in 3DNA/DSSR based on atomic names and planar geometry
Early versions of 3DNA employed only three atoms (N1, C2 and C6) and three distances to decide a nt. Purines were further discriminated by the N9 atom, and the N1–N9 distance. While developing DSSR, I revised the nt-identification algorithm by using a least-squares fitting procedure that makes use of all available base ring atoms instead of selected ones. The same new algorithm has also been adapted into the find_pair/analyze etc programs in 3DNA, as of v2.2.
As always, the idea can be best illustrated with a worked example. Guanine in its standard base reference frame, with the following list of nine ring atoms coordinates, is chosen for the least-squares fitting. See file Atomic_G.pdb in the 3DNA distribution, and also Table 1 of the report A Standard Reference Frame for the Description of Nucleic Acid Base-pair Geometry.
ATOM 2 N9 G A 1 -1.289 4.551 0.000
ATOM 3 C8 G A 1 0.023 4.962 0.000
ATOM 4 N7 G A 1 0.870 3.969 0.000
ATOM 5 C5 G A 1 0.071 2.833 0.000
ATOM 6 C6 G A 1 0.424 1.460 0.000
ATOM 8 N1 G A 1 -0.700 0.641 0.000
ATOM 9 C2 G A 1 -1.999 1.087 0.000
ATOM 11 N3 G A 1 -2.342 2.364 0.001
ATOM 12 C4 G A 1 -1.265 3.177 0.000
By using a ls-fitting procedure, only (any) three atoms are needed. We no longer need to make explicit selection, as we did previously (N1,C2,C6 and N9), thus allowing for possible modification on these atoms.
Using four nts (G1, 2MG10, H2U16, and PSU39, see Fig. 1 above top) of 1ehz as examples, the following list gives the atomic coordinates of base ring atoms, and root-mean-squres devisions (rmsd) of the least-squares fit. Of course, when performing least-squares fitting, the names of corresponding atoms must match (note the different ordering of atoms for H2U and PSU in the list vs the above standard G reference).
#G1, rmsd=0.008
ATOM 14 N9 G A 1 51.628 45.992 53.798 1.00 93.67 N
ATOM 15 C8 G A 1 51.064 46.007 52.547 1.00 92.60 C
ATOM 16 N7 G A 1 51.379 44.966 51.831 1.00 91.19 N
ATOM 17 C5 G A 1 52.197 44.218 52.658 1.00 91.47 C
ATOM 18 C6 G A 1 52.848 42.992 52.425 1.00 90.68 C
ATOM 20 N1 G A 1 53.588 42.588 53.534 1.00 90.71 N
ATOM 21 C2 G A 1 53.685 43.282 54.716 1.00 91.21 C
ATOM 23 N3 G A 1 53.077 44.429 54.946 1.00 91.92 N
ATOM 24 C4 G A 1 52.356 44.836 53.879 1.00 92.62 C
#2MG10, rmsd=0.018
HETATM 207 N9 2MG A 10 61.581 47.402 18.752 1.00 42.14 N
HETATM 208 C8 2MG A 10 62.199 48.621 18.635 1.00 40.38 C
HETATM 209 N7 2MG A 10 63.494 48.534 18.422 1.00 40.70 N
HETATM 210 C5 2MG A 10 63.745 47.167 18.395 1.00 43.82 C
HETATM 211 C6 2MG A 10 64.965 46.449 18.205 1.00 43.45 C
HETATM 213 N1 2MG A 10 64.767 45.086 18.293 1.00 44.71 N
HETATM 214 C2 2MG A 10 63.541 44.482 18.486 1.00 47.21 C
HETATM 217 N3 2MG A 10 62.411 45.125 18.614 1.00 45.85 N
HETATM 218 C4 2MG A 10 62.574 46.451 18.582 1.00 43.27 C
#H2U16, rmsd=0.188
HETATM 336 N1 H2U A 16 77.347 53.323 34.582 1.00 91.19 N
HETATM 337 C2 H2U A 16 76.119 52.865 34.160 1.00 92.39 C
HETATM 339 N3 H2U A 16 75.123 52.894 35.107 1.00 93.28 N
HETATM 340 C4 H2U A 16 75.289 52.711 36.458 1.00 93.34 C
HETATM 342 C5 H2U A 16 76.696 52.479 36.909 1.00 93.77 C
HETATM 343 C6 H2U A 16 77.717 53.238 36.039 1.00 93.22 C
#PSU39, rmsd=0.004
HETATM 845 N1 PSU A 39 74.080 36.066 5.459 1.00 75.82 N
HETATM 846 C2 PSU A 39 74.415 36.835 4.354 1.00 75.59 C
HETATM 847 N3 PSU A 39 75.735 36.769 3.984 1.00 76.29 N
HETATM 848 C4 PSU A 39 76.728 36.038 4.591 1.00 77.28 C
HETATM 849 C5 PSU A 39 76.307 35.280 5.732 1.00 77.93 C
HETATM 850 C6 PSU A 39 75.025 35.316 6.112 1.00 76.07 C
As noted in the DSSR paper, the rmsd is normally <0.1 Å since base rings are rigid. To account for experimental error and special non-planar cases, such as H2U in 1ehz, the default rmsd cutoff is set to 0.28 Å by default.
With the above detailed algorithm, DSSR (and the 3DNA find_pair/analyze programs) can automatically identify virtually all ‘recognizable’ nts in the PDB. A survey performed in June 2015 detected 630 different types of modified nucleotides in the PDB.
It is worth noting the following points:
- The choice of standard G instead of A as the reference base has no impact on the results. As a matter of fact, the rmsd between G and A is only 0.04 Å. Note also the generous default cutoff of 0.28 Å.
- The method obviously depends on proper naming of the ring atoms. Specially, the base ring atoms must be named
N1,C2,N3,C4,C5,C6 consecutively, with purines having three additional atoms named N7,C8,N9. Thus, under this scheme, TPP (thiamine diphosphate) would not be recognized as a nt by default, simply because of the extra prime (′) of atoms in the six-membered ring. In nucleic acid structures, the prime symbol is normally associated with atoms of the sugar moiety (e.g., the C5′ atom).
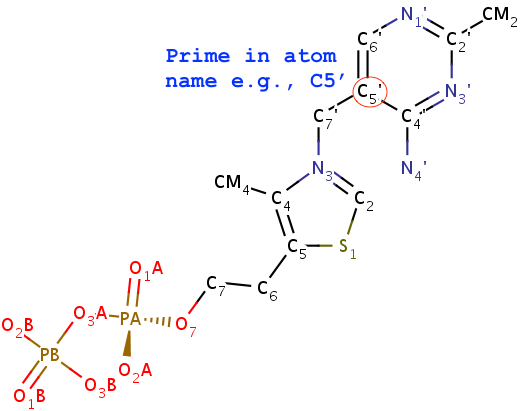
Fig. 3: TPP (thiamine diphosphate) would not be recognized as a nt.
- On the other hand, nt cofactors in an otherwise ‘pure’ protein structure will also be recognized. One example is the two AMP (adenosine monophosphate) ligands in PDB entry 12as. This extra identification of nts does no harm in such cases. As shown in the analysis of the SAM-I riboswitch in the DSSR paper, taking the SAM ligand as a nt in base triplet recognition is a neat feature.
- Once a nucleotide has been identified and classified into purines and pyrimidines, exocyclic atoms can be used for further assignment:
O6 or N2 distinguishes guanine from adenine, N4 separates cytosine from thymine and uracil, and C7 (or C5M, the methyl group) differentiates thymine from uracil. For some modified nts, the distinctions within purines or pyrimidines may not be that obvious. For example, inosine may be taken as a modified guanine or adenine. However, this ambiguity does not pose any significant effect on the calculated base-pair parameters.
- In DSSR and 3DNA, each identified nt is assigned a one-letter shorthand code: the standard
..A, .DA, and ADE (among a few other common variations) is shortened to upper-case A, and similarly for C, G, T, and U. Modified nts, on the other hand, are shortened to their corresponding lower-case symbol. For example, modified guanine such as 2MG and M2G in the yeast phenylalanine tRNA (see Fig. 1 above) is assigned g. So in 3DNA/DSSR output, the upper and lower cases of bases (e.g., nts=3 gCG A.2MG10,A.C25,A.G45) convey special meanings.
Related topics:

In DSSR (and find_pair -p from the original 3DNA suite), multiplets is defined as “three or more bases associated in a coplanar geometry via a network of hydrogen-bonding interactions. Multiplets are identified through inter-connected base pairs, filtered by pair-wise stacking interactions and vertical separations to ensure overall coplanarity.”
DSSR detects multiplets automatically, and outputs a corresponding MODEL/ENDMDL delineated PDB file (dssr-multiplets.pdb by default) where each multiplet is laid in the most extended view in terms of base planes. The DSSR Nucleic Acids Research (NAR) paper contains four examples (in supplemental Figures 1, 3, 4, and 7) to illustrate this functionality. Please refer to Reproducing results published in the DSSR-NAR paper on the 3DNA Forum for details.
Recently, I read the article titled InterRNA: a database of base interactions in RNA structures by Appasamy et al. in NAR. In Figure 2 of the paper, the authors showcased a sextuple (hexaplet) identified in the E. coli ribosome (PDB id: 4tpe), along with six base-base H-bonds contained therein.
With interest, I tried to run DSSR on the PDB entry 4tpe. As it turns out, ‘4tpe’ has been merged into 4u27 in mmCIF format. I ran DSSR (v1.4.6-2015dec16) in its default settings on ‘4u27’ and get the following summary of results.
# x3dna-dssr -i=4u27.cif -o=4u27.out
total number of base pairs: 4822
total number of multiplets: 680
total number of helices: 264
total number of stems: 566
total number of isolated WC/wobble pairs: 193
total number of atom-base capping interactions: 615
total number of hairpin loops: 215
total number of bulges: 137
total number of internal loops: 244
total number of junctions: 108
total number of non-loop single-stranded segments: 83
total number of kissing loops: 14
total number of A-minor (type I and II) motifs: 246
total number of ribose zippers: 127
total number of kink turns: 15
Among the 680 DSSR-identified multiplets, two hexaplets (one on chain “AA”, and another on “CA”) match those reported by Appasamy et al., as shown below:
678 nts=6 GUUAAA 1:AA.G404,1:AA.U438,1:AA.U439,1:AA.A496,1:AA.A498,1:AA.A499
679 nts=6 GUUAAA 1:CA.G404,1:CA.U438,1:CA.U439,1:CA.A496,1:CA.A498,1:CA.A499
For illustration, the hexaplet #678 is extracted from dssr-multiplets.pdb to file 4u27-hexaplet.pdb (download the coordinates) and shown below. The figure is generated by DSSR and PyMOL, as detailed in Reproducing results published in the DSSR-NAR paper on the 3DNA Forum.
x3dna-dssr -i=4u27-hexaplet.pdb -o=4u27-hexaplet.pml --hbfile-pymol
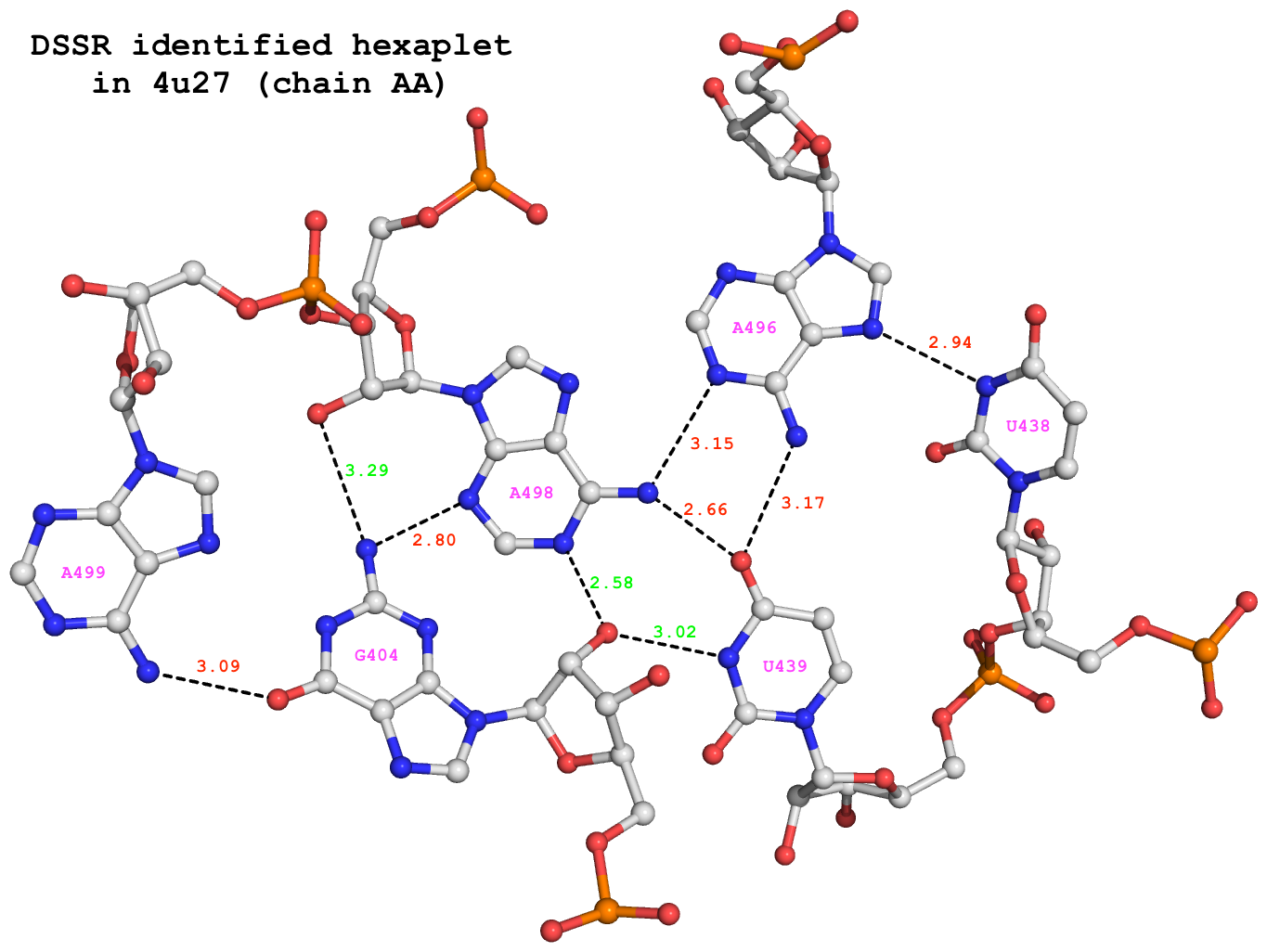
DSSR-identified hexaplet GUUAAA in 4u27.
DSSR identifies 6 base pairs in the hexaplet:
# x3dna-dssr -i=4u27-hexaplet.pdb --idstr=short
List of 6 base pairs
nt1 nt2 bp name Saenger LW DSSR
1 G404 A498 G+A -- n/a tSS tm+m
2 G404 A499 G+A -- n/a cWH cW+M
3 U438 A496 U-A rHoogsteen 24-XXIV tWH tW-M
4 U439 A496 U-A -- n/a cH. cM-.
5 U439 A498 U-A WC 20-XX cWW cW-W
6 A496 A498 A+A -- n/a cWH cW+M
It detects a total of 9 H-bonds as shown below. In addition to the 6 base-base H-bonds noted by Appasamy et al., DSSR also finds 3 sugar-base H-bonds (#1, #2, and #4, labeled in green) that obviously play a role in stabilizing the high-order base association.
# x3dna-dssr -i=4u27-hexaplet.pdb --get-hbonds --idstr=short
11 59 #1 o 3.017 O:N O2'@G404 N3@U439
11 104 #2 o 2.578 O:N O2'@G404 N1@A498
18 125 #3 p 3.089 O:N O6@G404 N6@A499
21 96 #4 o 3.289 N:O N2@G404 O2'@A498
21 106 #5 p 2.797 N:N N2@G404 N3@A498
39 78 #6 p 2.944 N:N N3@U438 N7@A496
61 81 #7 p 3.167 O:N O4@U439 N6@A496
61 103 #8 p 2.662 O:N O4@U439 N6@A498
82 103 #9 p 3.152 N:N N1@A496 N6@A498

From the Jmol mailing list, I noticed Jmol 14.4.0 was released yesterday (October 13, 2015) by Dr. Bob Hanson. Among the development highlights is the following item:
biomolecule annotations including DSSR, RNA3D, EBI sequence domains, and PDB validation data
I am glad to see that DSSR has been integrated into Jmol, one of the most popular molecular graphics visualization programs. To enable easy access to the DSSR functionality from Jmol, I’ve set up two websites with easy-to-remember URLs: http://jmol.x3dna.org and http://jsmol.x3dna.org. They both point to the same jsmol/ folder extracted from jsmol.zip of the Jmol distribution.
In retrospect, I first met Bob at the Workshop on the PDBx/mmCIF Data Exchange Format for Structural Biology held at Rutgers University during October 21-22, 2013. I approached him during a lunch break, asking for a possible collaboration on integrating DSSR into Jmol. The name DSSR may have played a role in convincing Bob, since it matches the well-known DSSP program for proteins. In the end, we were both excited about the project, talked into details after the meeting, and continued our conversation the next morning while I drove him to the airport.
Nothing real happened until early April 2014. Once getting started, however, we moved forward rapidly: it took less then three weeks to get the first functional version ready for the community to play. See Bob’s announcement RNA/DNA Secondary Structure, anyone? in the Jmol mailing list on April 9, 2014. During this process, we communicated extensively via email, up to 30 messages per day, on technical details for better communication between the two programs. The integration works by using Jmol as a front-end, which calls a web-serivce hosted at Columbia University for DSSR analysis. Jmol’s parsing of the DSSR output is facilitated by the dedicated --jmol option.
The above preliminary, yet functional, DSSR-Jmol integration had be in service without infrastructural changes until two months ago. In August 10, 2015, Bob contacted me:
I might make a significant request though. That would be for the server to deliver all this in JSON format. This is really the way to go. It is what people want and it is perfect for Jmol as well.
I’d played around with JSON or SQLite as a structured data exchange format for quite some time, and Bob’s request finally convinced me that JSON is the (better) way to go. And that began another around of intensive collaborative work that has switched the exchange format between DSSR and Jmol from plain text output to JSON. From August 10 to September 22, we had a total of over 170-email exchanges, plus Skype. JSON has really simplified lives of both parties, especially in the long run.
Overall, collaborating with Bob has been truly an enjoyable and rewarding experience. The DSSR-Jmol integration also serves as a concrete example of what can be achieved by two dedicated minds with complementary expertise.

Over the past couple of weeks, I’ve added two more DSSR options, --symmetry and --nmr, that are closely related to an ensemble of MODEL/ENDMDL-delineated structures in PDB files. However, there exist subtle differences between the two cases, and the usage of the same MODEL/ENDMDL ensemble format can be ambiguous to the uninitiated. This blog post aims to clarify the issues, using concrete examples.
The --symmetry options applies to X-ray crystal structures where an asymmetric unit represents only part of the whole biological assembly. In standard PDB format, the asymmetric unit contains instructions to produce crystallographic symmetry
related molecules.. Nevertheless, the biological assembly are also provided by the PDB (or NDB), with coordinate files ending with .pdb1 or such. For example, the PDB entry 2d94 has the single-stranded sequence GGGCGCCC in its asymmetric unit (2d94.pdb). It is the biological assembly in file 2d94.pdb1 that contains the DNA double helix.
x3dna-dssr -i=2d94.pdb # no pairs found
x3dna-dssr -i=2d94.pdb1 # still no pairs found
x3dna-dssr -i=2d94.pdb1 --symm # 8 pairs found
x3dna-dssr -i=2d94.pdb --symm # no pairs found
As shown by the above examples, DSSR by default reads only the first model even given the biological assemble file 2d94.pdb1. It is with --symmetry (abbreviated to --symm) explicitly specified that DSSR takes all models in the input biological assemble file into consideration. The last case also illustrates that DSSR does not generate crystallographic symmetry related molecules. The --symm simply informs DSSR to take all models, which already exist in the input file, into consideration.
On the other hand, the --nmr option is for auto-processing an ensemble of structures solved by solution NMR method (or trajectories of molecular dynamics simulations). The key point here is that each of the MODEL/ENDMDL-delinated structures is independent and thus can be processed separately, even though they are obviously closely related. Using the PDB entry 2n2d as an example, here are some sample usages:
x3dna-dssr -i=2n2d.pdb -o= 2n2d-first.out # only the first structure is processed
x3dna-dssr -i=2n2d.pdb --nmr -o=2n2d-all.out # all 10 structures are processed
x3dna-dssr -i=2n2d.pdb --nmr --json -o=2n2d-all.json # ibid., with output in JSON
Note that the NMR file is named 2n2d.pdb, and it contains 10 structures.
Interesting mixes show up when an X-ray biological assembly with multiple MODEL/ENDMDL entries is analyzed with --nmr, or an NMR entry is handled with --symmetry. Here are two such examples:
x3dna-dssr -i=2d94.pdb1 --nmr -o=temp # models 1 and 2 are handled sepatately
x3dna-dssr -i=2n2d.pdb --symm -o=temp # wrong -- does not make sense!
In summary, the --symmetry option is intended to treat symmetry-related molecules as a whole, as in a biological assembly of X-ray crystal structures. In contrast, the --nmr option aims to automate the analysis of each structure in a MODEL/ENDMDL-delineated ensemble, as in NMR structures or trajectories of MD simulations. The distinction between the two MODEL/ENDMDL usages is most clearly seen via a molecular visualization program: for example, check the figure below for 2d94.pdb1 (left) and 2n2d.pdb (right) when all frames are selected using Jmol.

Recently, I read with great interest an article titled A context-sensitive guide to RNA & DNA base-pair & base-stack geometry by Dr. Jane Richardson, published in CCN (Computational Crystallography Newsletter, 2015, 5, 42—49). Highlighted in the article are Buckle and Propeller twist (see bottom left of the figure below), two of the angular parameters that characterize base-pair (bp) non-planarity. Particularly, I was intrigued by the “Notes on measures and figures” at the end:
Base normals were constructed in Mage (Richardson 2001) and twist torsions and buckle angles were measured from them; propeller-twists were measured as dihedral angles around an axis between N1/9 atoms.

The Richardson CCN article prompted me to think more on intuitive description of bp geometry that can be easily grasped by experimentalist, especially X-ray crystallographers or cryo-EM practitioners. Without worrying about model building as with the six rigid-body parameters, it is straightforward to come up with a new set of four ‘simple’ parameters (Shear, Stretch, Buckle and Propeller) with the following characteristics:
- Each parameter can be positive or negative. For type M–N pairs (as in the canonical cases), Shear and Buckle reverse their signs when the two bases are swapped (i.e. counted as N–M instead of M–N). In all other cases, the signs of the parameters remain unchanged. See the DSSR paper for the definition of M+N vs M–N type of pairs.
- Intuitive results for non-canonical pairs, even when Opening is ~180º.
- Consistent definition between Shear/Buckle (_x_-axis) vs Stretch/Propeller (_y_-axis).
- As in 3DNA and DSSR, Buckle^2 + Propeller^2 = interBase_angle^2. Either Buckle or Propeller can render the two base planes of a pair non-parallel. Combined together, they introduce a non-zero inter-base angle. By definition, each parameter should not be larger than the overall inter-base angle.
With the cartoon-block representation introduced in DSSR, base-stacking interactions and bp deformations (especially Buckle and Propeller) are immediately obvious. Two example are illustrated in the figure below: one is the classic Dickerson B-DNA dodecamer (355d, DSSR output), and the other is the parallel double-stranded helix of poly(A) RNA (4jrd, DSSR output).
A portion of DSSR output for the B-DNA duplex 355d is shown below. Note that the first bp (at the bottom left in the figure above) has a Propeller of –17º (and a Buckle of +7º). As beautifully explained by Calladine et al. in their book Understanding DNA,
The Molecule & How It Works, Watson-Crick pairs prefer to have negative Propeller in right-handed DNA double helices to improve same-strand base-stacking interactions. The average value of Propeller in A- and B-DNA crystal structures is around –11º (see Table 3 of the Olson et al. standard base reference frame paper).
nt1 nt2 bp name Saenger LW DSSR
1 A.DC1 B.DG24 C-G WC 19-XIX cWW cW-W
[-105.9(anti) ~C2'-endo lambda=53.5] [-141.3(anti) ~C3'-endo lambda=52.7]
d(C1'-C1')=10.71 d(N1-N9)=8.96 d(C6-C8)=9.88 tor(C1'-N1-N9-C1')=-21.4
H-bonds[3]: "O2(carbonyl)-N2(amino)[2.83],N3-N1(imino)[2.90],N4(amino)-O6(carbonyl)[2.98]"
interBase-angle=19 Simple-bpParams: Shear=0.28 Stretch=-0.13 Buckle=7.3 Propeller=-17.2
bp-pars: [0.28 -0.14 0.07 6.93 -17.31 -0.61]
2 A.DG2 B.DC23 G-C WC 19-XIX cWW cW-W
[-85.4(anti) ~C2'-endo lambda=53.4] [-150.3(anti) ~C3'-endo lambda=55.4]
d(C1'-C1')=10.61 d(N1-N9)=8.92 d(C6-C8)=9.83 tor(C1'-N1-N9-C1')=-21.7
H-bonds[3]: "O6(carbonyl)-N4(amino)[2.91],N1(imino)-N3[2.88],N2(amino)-O2(carbonyl)[2.88]"
interBase-angle=17 Simple-bpParams: Shear=-0.24 Stretch=-0.18 Buckle=9.0 Propeller=-14.5
bp-pars: [-0.24 -0.18 0.49 9.34 -14.30 -2.08]
A portion of DSSR output for the parallel A-DNA duplex 4jrd is shown below. Note that the values of ‘simple’ Propeller are positive for both bps #7 and #8. In contrast, the rigid-body bp parameters have their signs flipped over when Opening is switched from –179.56º for bp#7 to +179.23º for bp#8. This sign ‘ambiguity’ around 180º Opening could be confusing. Yet, all the six bp parameters must be kept as they are for rigorous rebuilding, especially within a larger context than a bp per se. From the very beginning, 3DNA has adopted the convention of keeping angular parameters in the range of [–180º, +180º] instead of [0, 360º], allowing left-handed Z-DNA to have negative twist.
7 A.A8 B.A7 A+A -- 02-II tHH tM+M
[-175.8(anti) ~C3'-endo lambda=10.2] [-172.7(anti) ~C3'-endo lambda=12.6]
d(C1'-C1')=11.15 d(N1-N9)=8.29 d(C6-C8)=6.31 tor(C1'-N1-N9-C1')=160.1
H-bonds[4]: "OP2-N6(amino)[2.97],N7-N6(amino)[2.97],N6(amino)-OP2[2.92],N6(amino)-N7[2.91]"
interBase-angle=14 Simple-bpParams: Shear=-7.88 Stretch=0.66 Buckle=-7.8 Propeller=11.9
bp-pars: [-6.00 5.15 -0.02 0.63 14.22 -179.56]
8 A.A9 B.A8 A+A -- 02-II tHH tM+M
[-177.4(anti) ~C3'-endo lambda=12.4] [-175.8(anti) ~C3'-endo lambda=10.3]
d(C1'-C1')=11.01 d(N1-N9)=8.15 d(C6-C8)=6.18 tor(C1'-N1-N9-C1')=158.5
H-bonds[4]: "OP2-N6(amino)[2.93],N7-N6(amino)[2.88],N6(amino)-OP2[2.97],N6(amino)-N7[2.92]"
interBase-angle=15 Simple-bpParams: Shear=-7.91 Stretch=0.56 Buckle=-7.0 Propeller=13.7
bp-pars: [6.11 -5.06 -0.05 -2.26 -15.22 179.23]

Standard nitrogenous bases in DNA and RNA (A, C, G, T, and U) are aromatic compounds, each with a planar geometry. In the analyses of three-dimensional (3D) nucleic acid structures, the planar bases are normally taken as rigid bodies. The relative geometry of the two bases in base pair (bp) can then be rigorously quantified by six rigid-body parameters (see figure below). The three translations along the _x_-, _y_- and _z_-axes are termed Shear, Stretch, and Stagger, respectively. The three corresponding rotations are called Buckle, Propeller (twist), and Opening.

3DNA is unique with its coupled analyze and rebuild programs. The former calculates six bp parameters given 3D atomic coordinates (in PDB or PDBx/mmCIF format), while the later takes a set of such parameters to generate the corresponding structure. The rigor of the description can be easily verified in two equivalent ways: the close to zero root-mean-square deviation (RMSD) between the rebuilt structure and the original coordinates, after a least-squares superposition; or the identical six bp parameters when the rebuilt structure is analyzed.
As is often the case, a concrete example would make the point clear. Here I am using the reverse Hoogsteen (rHoogsteen) bp between U8 and A14 (see image below) in the yeast phenylalanine tRNA (1ehz) as an example. The PDB atomic coordinates of the U8–A14 rHoogsteen pair, excluding backbone atoms except for C1′, is stored in file 1ehz-U8-A14.pdb.
find_pair 1ehz-U8-A14.pdb stdout | analyze stdin
# bp parameters in file '1ehz-U8-A14.out'
# also generated 'bp_step.par' for rebuilding below
rebuild -atomic bp_step.par 1ehz-U8-A14-3DNA.pdb
# rmsd is 0.044 Å between '1ehz-U8-A14.pdb' and '1ehz-U8-A14-3DNA.pdb'
find_pair 1ehz-U8-A14-3DNA.pdb stdout | analyze stdin
# bp parameters of the rebuilt structure in '1ehz-U8-A14-3DNA.out'
rebuild -atomic bp_step.par 1ehz-U8-A14-3DNA-new.pdb
# rmsd is 0 Å between '1ehz-U8-A14-3DNA.pdb' and '1ehz-U8-A14-3DNA-new.pdb'
Note that the above commands should be performed in order, since the file bp_step.par is overwritten after each analyze run. For your verification, here are the links to the five files:
The 0.044 Å rmsd between the original PDB coordinates in 1ehz-U8-A14.pdb and the 3DNA rebuilt structure in 1ehz-U8-A14-3DNA.pdb is due to the slight non-planarity of experimental bases. The rmsd is 0 between the two rounds of 3DNA rebuilt structures, 1ehz-U8-A14-3DNA.pdb and 1ehz-U8-A14-3DNA-new.pdb, as expected.
The bp parameters in 1ehz-U8-A14.out and 1ehz-U8-A14-3DNA.out are identical, as expected, and they are shown below.
Local base-pair parameters
bp Shear Stretch Stagger Buckle Propeller Opening
1 U-A 4.14 -1.91 0.77 -4.62 12.12 -103.09
Running DSSR on 1ehz-U8-A14.pdb gives the following results. Note that the six bp parameters (last row prefixed with bp-pars) are the exactly same as in 3DNA — we are consistent.
# x3dna-dssr -i=1ehz-U8-A14.pdb --more
List of 1 base pair
nt1 nt2 bp name Saenger LW DSSR
1 A.U8 A.A14 U-A rHoogsteen 24-XXIV tWH tW-M
[n/a(n/a) ---- lambda=28.3] [n/a(n/a) ---- lambda=21.5]
d(C1'-C1')=9.63 d(N1-N9)=7.06 d(C6-C8)=6.00 tor(C1'-N1-N9-C1')=174.4
H-bonds[2]: "O2(carbonyl)-N6(amino)[3.00],N3(imino)-N7[2.74]"
interBase-angle=12.97 Simple-bpParams: Shear=4.28 Stretch=1.55 Buckle=-11.8 Propeller=5.4
bp-pars: [4.14 -1.91 0.77 -4.62 12.12 -103.09]
As mentioned in the recent DSSR paper:
As in 3DNA (6,7), DSSR takes advantage of the six standard base-pair parameters––three translations (Shear, Stretch, Stagger) and three rotations (Buckle, Propeller, Opening)––to quantify the relative spatial position and orientation of any two interacting bases rigorously. Among the six parameters, only Shear, Stretch, and Opening are critical for characterizing different types of pairs. Buckle, Propeller and Stagger, on the other hand, describe the nonplanarity of a given pair (6). By virtue of the definition of the standard base reference frame, Shear, Stretch, and Opening are all close to zero for Watson-Crick pairs. Moreover, every other type of pair has a set of characteristic parameters. For example, the wobble G–U pair is characterized by an average Shear of –2.2 Å, and the Hoogsteen A+U pair is distinguished by a Stretch of approximately –3.5 Å and an Opening of near 66º.
In a follow-up post, I will talk about the “simple” bp parameters (Simple-bpParams in the above DSSR output list) recently introduced into DSSR — stay tuned!