In DSSR (and find_pair -p from the original 3DNA suite), multiplets is defined as “three or more bases associated in a coplanar geometry via a network of hydrogen-bonding interactions. Multiplets are identified through inter-connected base pairs, filtered by pair-wise stacking interactions and vertical separations to ensure overall coplanarity.”
DSSR detects multiplets automatically, and outputs a corresponding MODEL/ENDMDL delineated PDB file (dssr-multiplets.pdb by default) where each multiplet is laid in the most extended view in terms of base planes. The DSSR Nucleic Acids Research (NAR) paper contains four examples (in supplemental Figures 1, 3, 4, and 7) to illustrate this functionality. Please refer to Reproducing results published in the DSSR-NAR paper on the 3DNA Forum for details.
Recently, I read the article titled InterRNA: a database of base interactions in RNA structures by Appasamy et al. in NAR. In Figure 2 of the paper, the authors showcased a sextuple (hexaplet) identified in the E. coli ribosome (PDB id: 4tpe), along with six base-base H-bonds contained therein.
With interest, I tried to run DSSR on the PDB entry 4tpe. As it turns out, ‘4tpe’ has been merged into 4u27 in mmCIF format. I ran DSSR (v1.4.6-2015dec16) in its default settings on ‘4u27’ and get the following summary of results.
# x3dna-dssr -i=4u27.cif -o=4u27.out
total number of base pairs: 4822
total number of multiplets: 680
total number of helices: 264
total number of stems: 566
total number of isolated WC/wobble pairs: 193
total number of atom-base capping interactions: 615
total number of hairpin loops: 215
total number of bulges: 137
total number of internal loops: 244
total number of junctions: 108
total number of non-loop single-stranded segments: 83
total number of kissing loops: 14
total number of A-minor (type I and II) motifs: 246
total number of ribose zippers: 127
total number of kink turns: 15
Among the 680 DSSR-identified multiplets, two hexaplets (one on chain “AA”, and another on “CA”) match those reported by Appasamy et al., as shown below:
678 nts=6 GUUAAA 1:AA.G404,1:AA.U438,1:AA.U439,1:AA.A496,1:AA.A498,1:AA.A499 679 nts=6 GUUAAA 1:CA.G404,1:CA.U438,1:CA.U439,1:CA.A496,1:CA.A498,1:CA.A499
For illustration, the hexaplet #678 is extracted from dssr-multiplets.pdb to file 4u27-hexaplet.pdb (download the coordinates) and shown below. The figure is generated by DSSR and PyMOL, as detailed in Reproducing results published in the DSSR-NAR paper on the 3DNA Forum.
x3dna-dssr -i=4u27-hexaplet.pdb -o=4u27-hexaplet.pml --hbfile-pymol
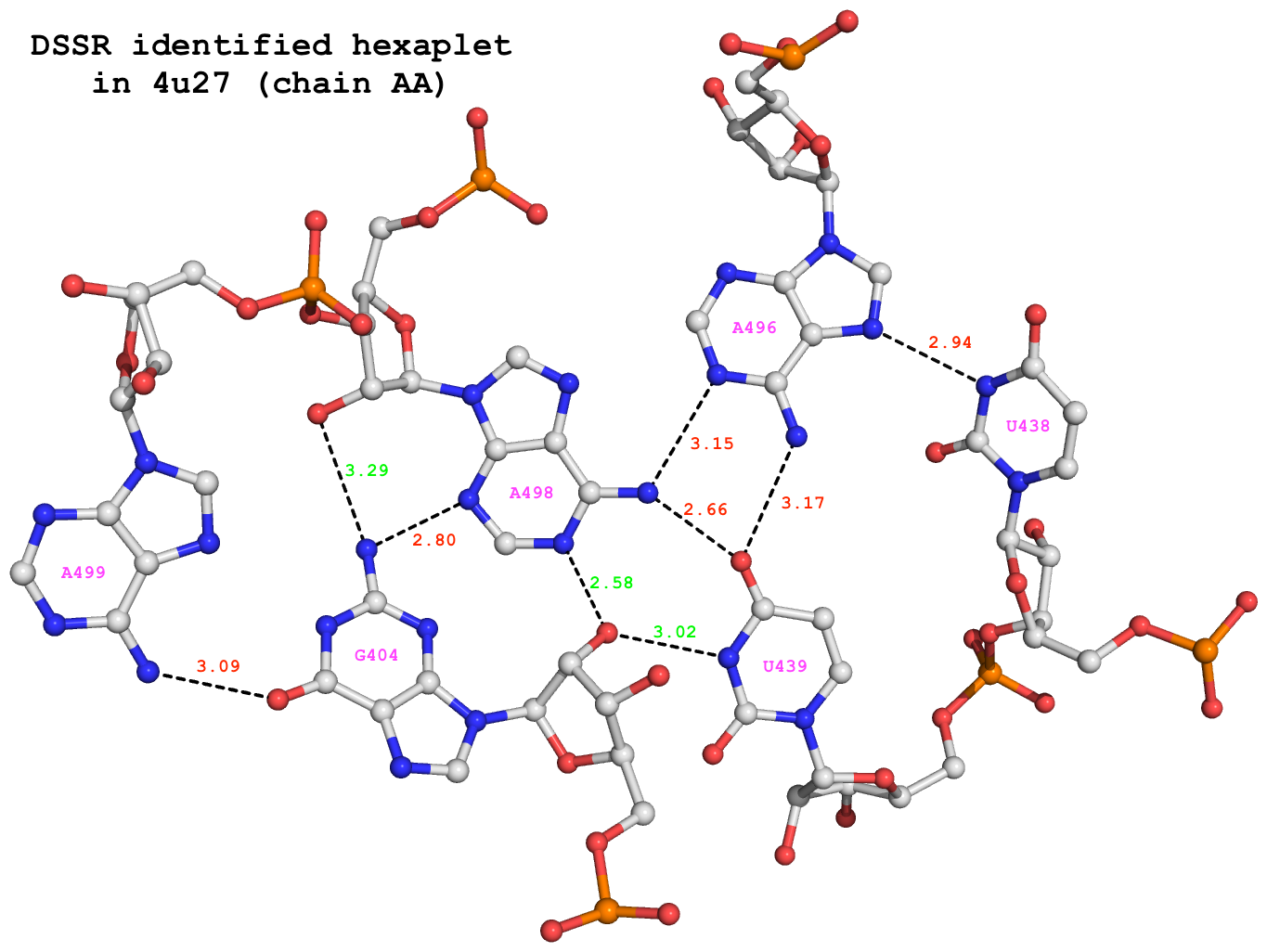
DSSR-identified hexaplet GUUAAA in 4u27.
DSSR identifies 6 base pairs in the hexaplet:
# x3dna-dssr -i=4u27-hexaplet.pdb --idstr=short
List of 6 base pairs
nt1 nt2 bp name Saenger LW DSSR
1 G404 A498 G+A -- n/a tSS tm+m
2 G404 A499 G+A -- n/a cWH cW+M
3 U438 A496 U-A rHoogsteen 24-XXIV tWH tW-M
4 U439 A496 U-A -- n/a cH. cM-.
5 U439 A498 U-A WC 20-XX cWW cW-W
6 A496 A498 A+A -- n/a cWH cW+M
It detects a total of 9 H-bonds as shown below. In addition to the 6 base-base H-bonds noted by Appasamy et al., DSSR also finds 3 sugar-base H-bonds (#1, #2, and #4, labeled in green) that obviously play a role in stabilizing the high-order base association.
# x3dna-dssr -i=4u27-hexaplet.pdb --get-hbonds --idstr=short 11 59 #1 o 3.017 O:N O2'@G404 N3@U439 11 104 #2 o 2.578 O:N O2'@G404 N1@A498 18 125 #3 p 3.089 O:N O6@G404 N6@A499 21 96 #4 o 3.289 N:O N2@G404 O2'@A498 21 106 #5 p 2.797 N:N N2@G404 N3@A498 39 78 #6 p 2.944 N:N N3@U438 N7@A496 61 81 #7 p 3.167 O:N O4@U439 N6@A496 61 103 #8 p 2.662 O:N O4@U439 N6@A498 82 103 #9 p 3.152 N:N N1@A496 N6@A498
