Ever since the 2003 publication of the initial 3DNA Nucleic Acids Research paper (NAR03), the schematic diagrams of base-pair parameters (see figure below) has become quite popular. Over the years, we have received numerous requests for permission to use the figure, or a portion thereof; as an example, the figure has been adopted into a structural biology textbook. In the 2008 3DNA Nature Protocols paper (NP08), we devoted the very first protocol to “create a schematic image for propeller of 45°”.
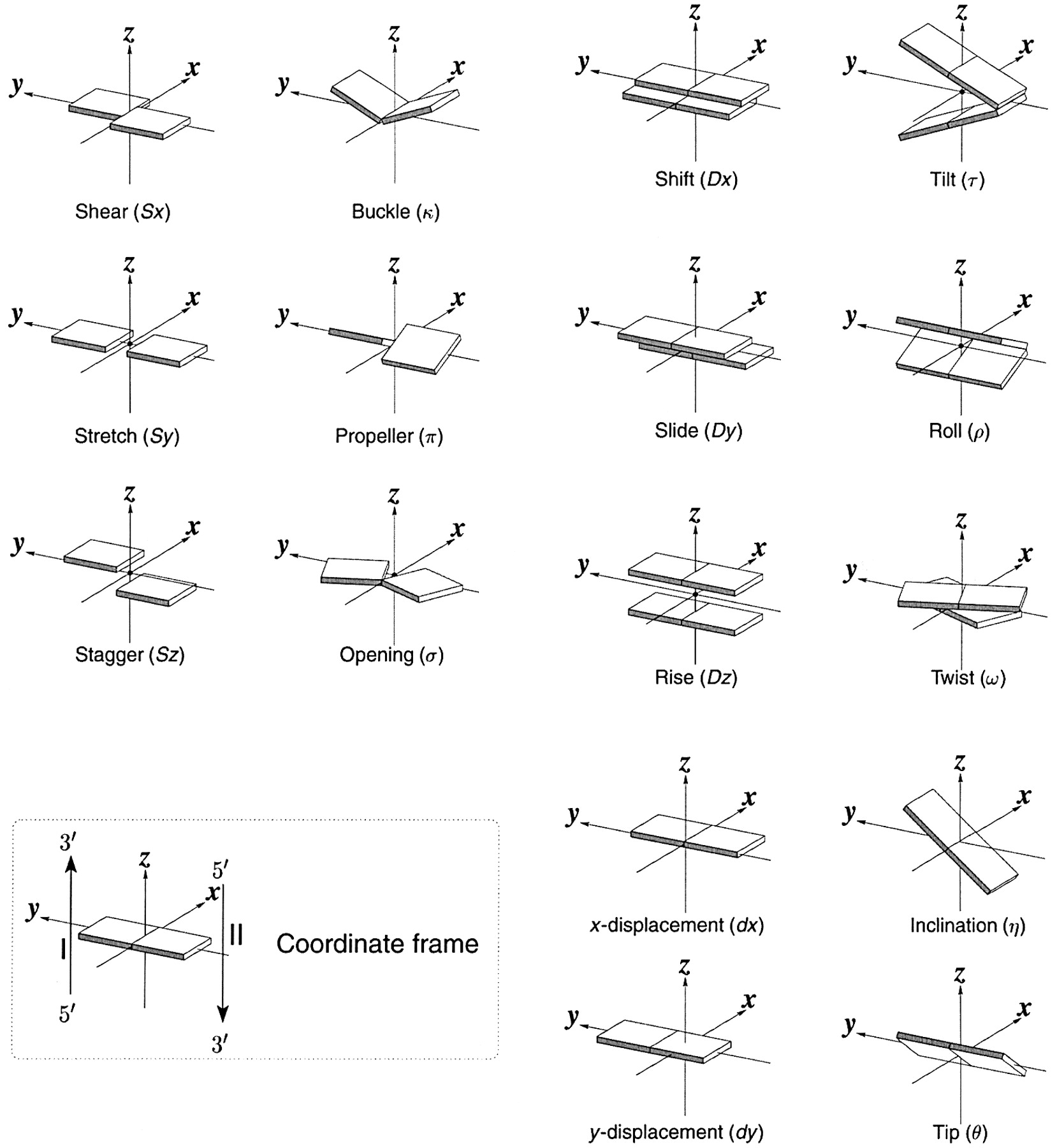
Figure legend taken from Figure 1 of NAR03: Pictorial definitions of rigid body parameters used to describe the geometry of complementary (or non‐complementary) base pairs and sequential base pair steps (19). The base pair reference frame (lower left) is constructed such that the x‐axis points away from the (shaded) minor groove edge of a base or base pair and the y‐axis points toward the sequence strand (I). The relative position and orientation of successive base pair planes are described with respect to both a dimer reference frame (upper right) and a local helical frame (lower right). Images illustrate positive values of the designated parameters. For illustration purposes, helical twist (Ωh) is the same as Twist (ω), formerly denoted by Ω (19,20) and helical rise (h) is the same as Rise (Dz).
I recall spending around two weeks to produce the above figure. Content-wise, the figure was constructed in only a short while; it was the little details that took me most of the time.
Over time, I’ve witnessed numerous versions of such schematic images in publications related to DNA/RNA structures. While looking similar, the schematics differ subtly in the magnitude, orientation and relative scale of illustrated parameters. To the best of my knowledge, only 3DNA provides a pragmatic approach to generate the base-pair schematic diagrams consistently.
To make the schematics more readily accessible, I’ve reproduced a high resolution image (in png format) for each of the 14 parameters shown above. You are welcome to pick and match the diagrams as necessary. If you use any of them in your publications, please cite the 3DNA NAR03 and/or NP08 paper(s).
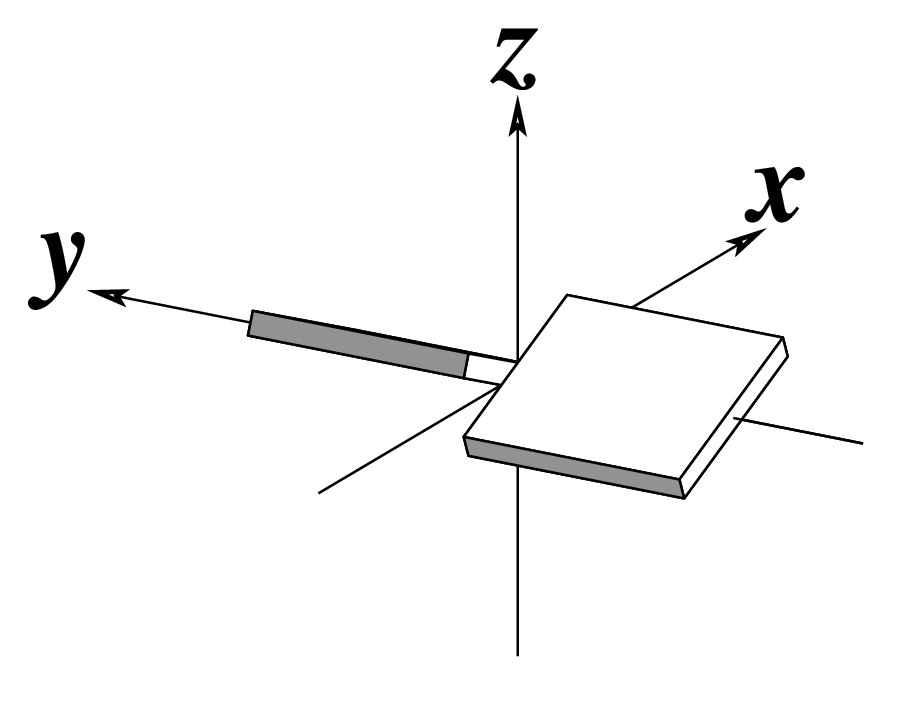
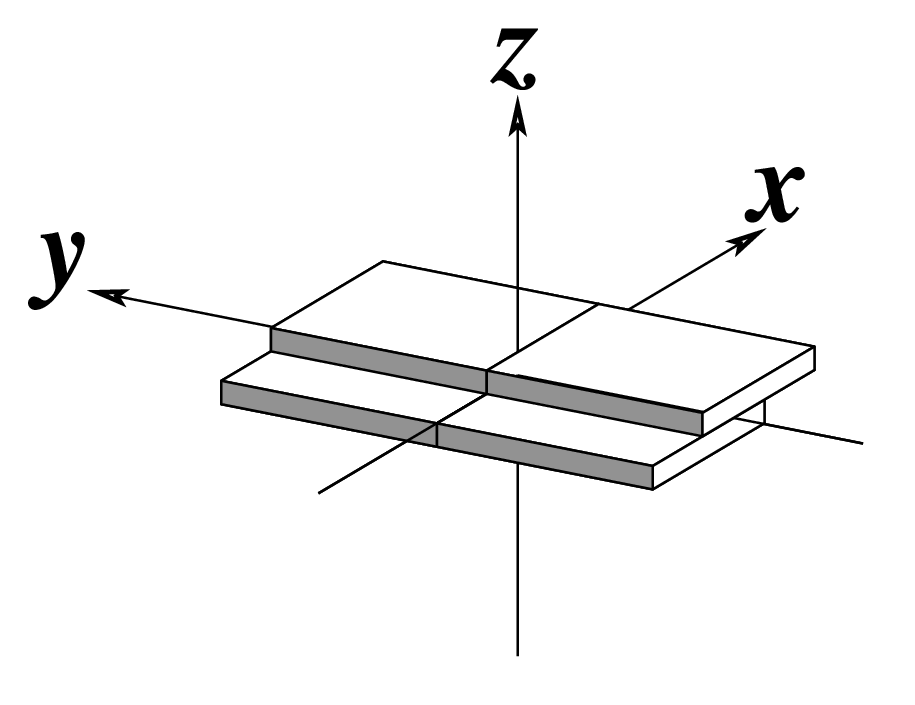
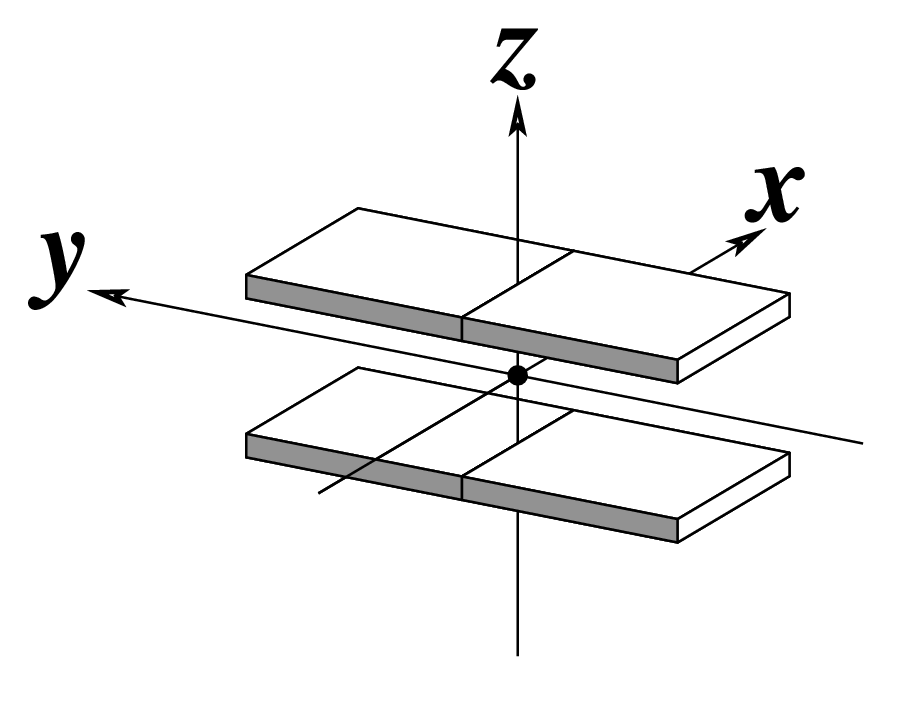
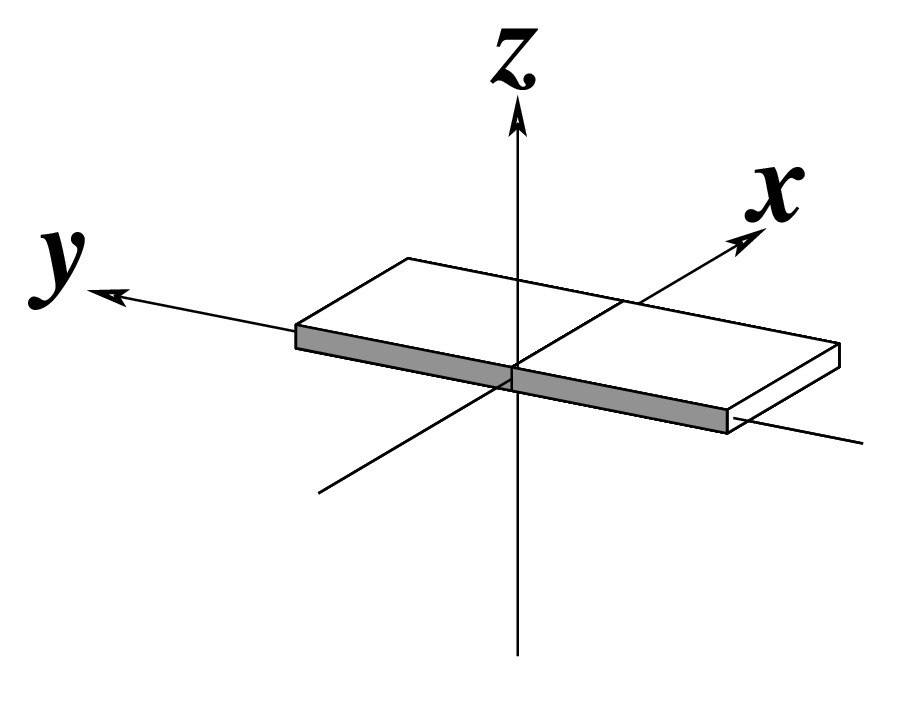
Note that in the schematic diagrams below, the shaded edge (facing the viewer) denotes the minor-groove side of a base or base pair.
| Shear (Sx) |
Stretch (Sy) |
Stagger (Sz) |
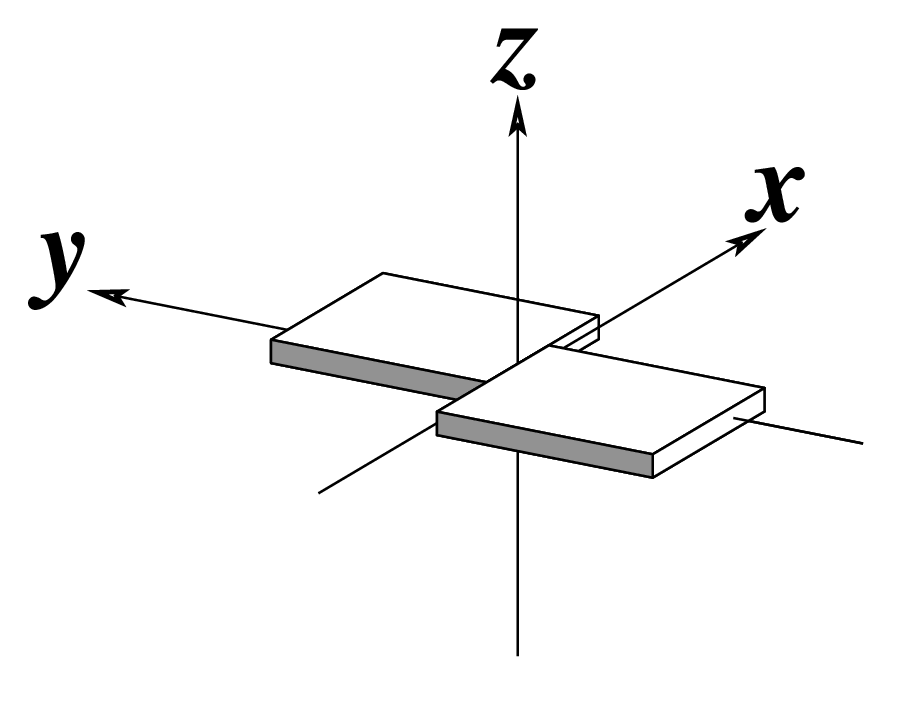 |
 |
 |
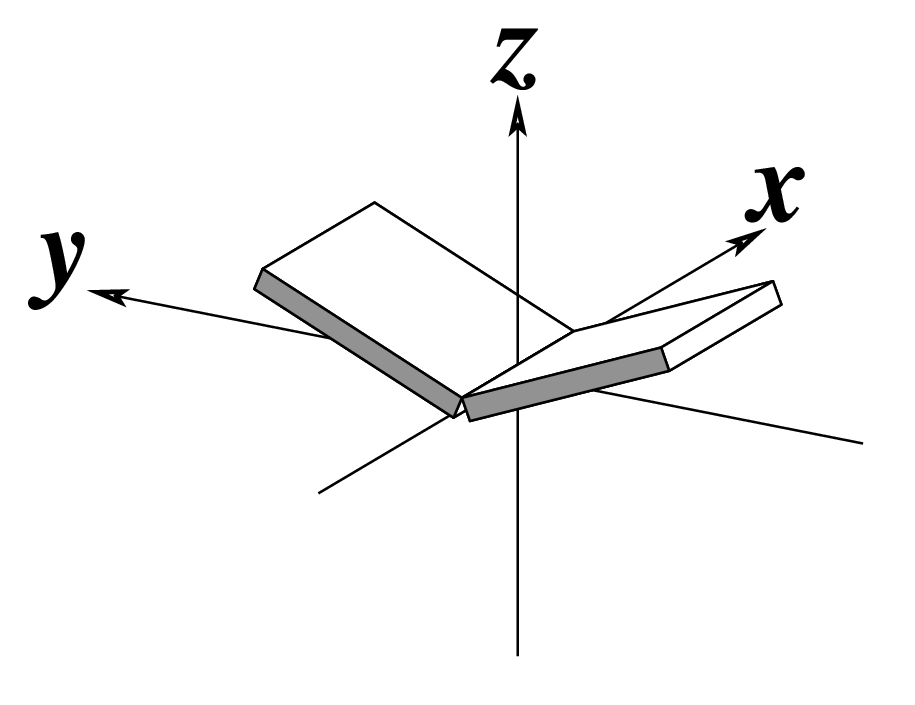
| Buckle (κ) |
Propeller (π) |
Opening (σ) |
 |
 |
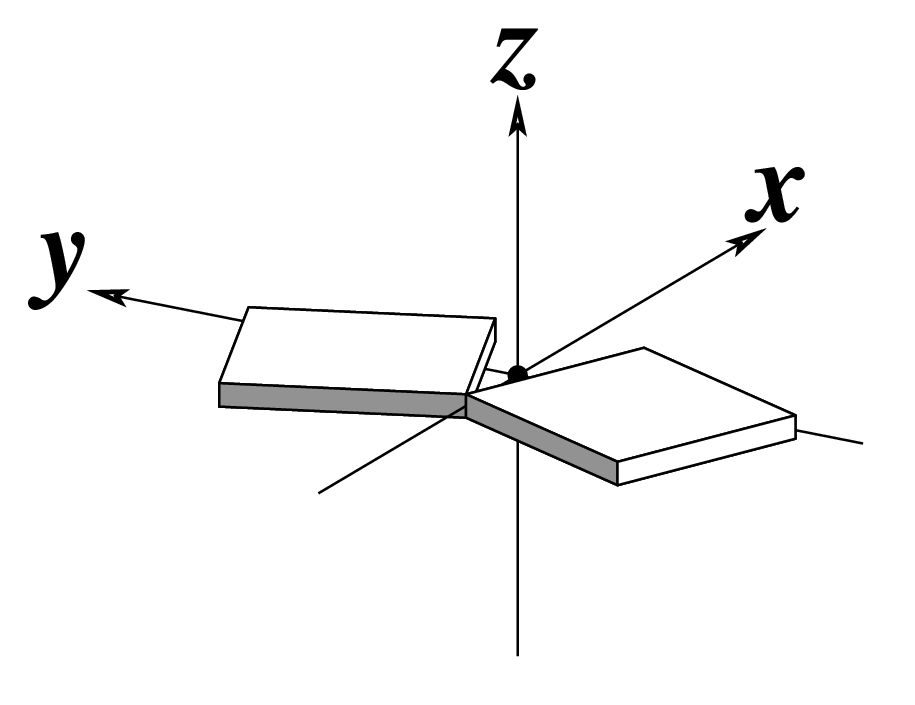 |
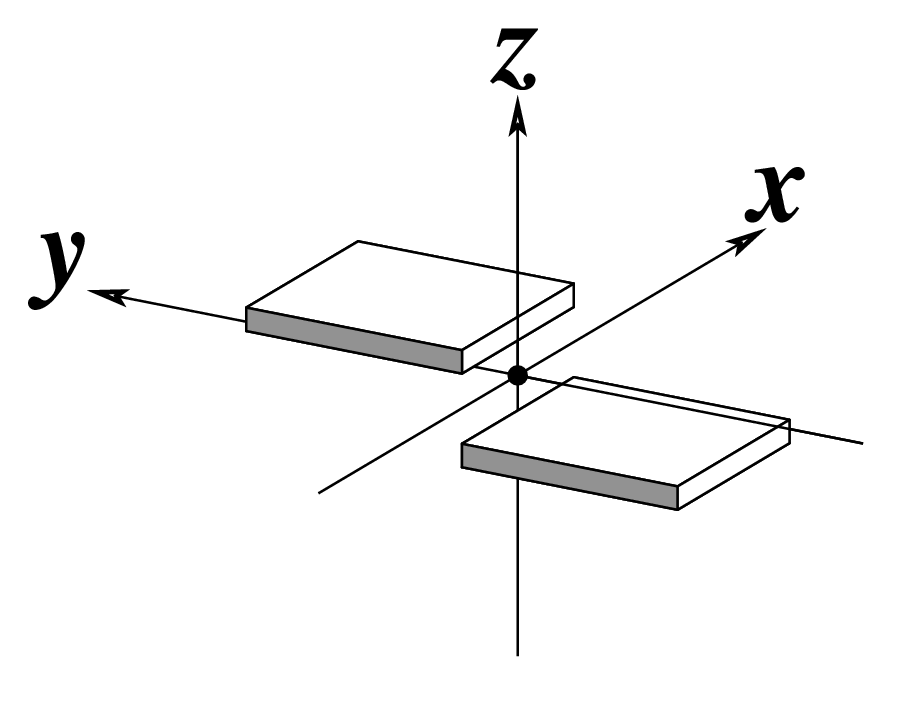
| Shift (Dx) |
Slide (Dy) |
Rise (Dz) |
 |
 |
 |
| Tilt (τ) |
Roll (ρ) |
Twist (ω) |
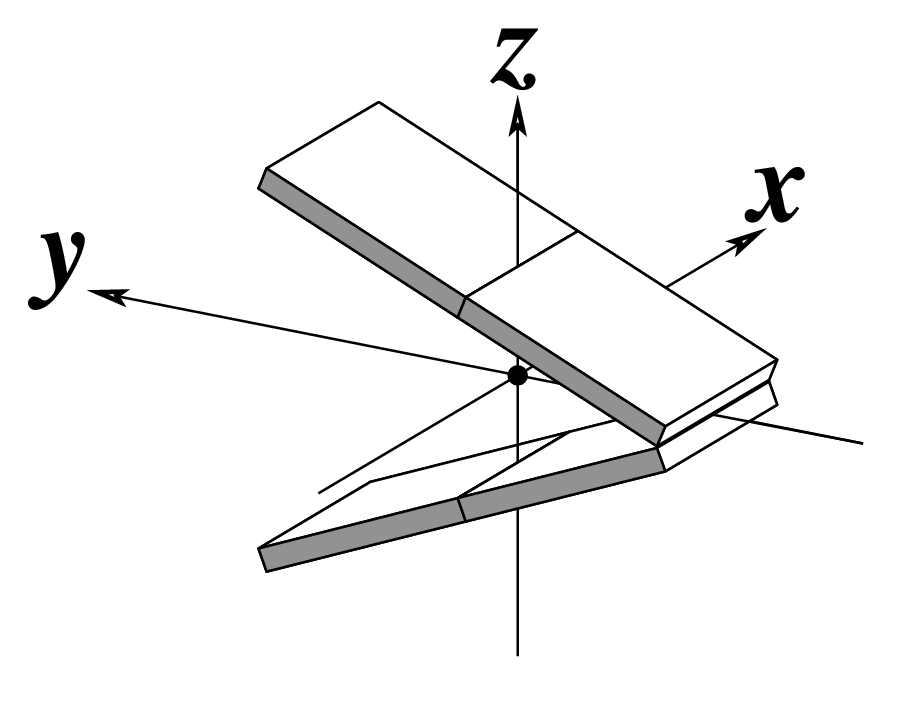 |
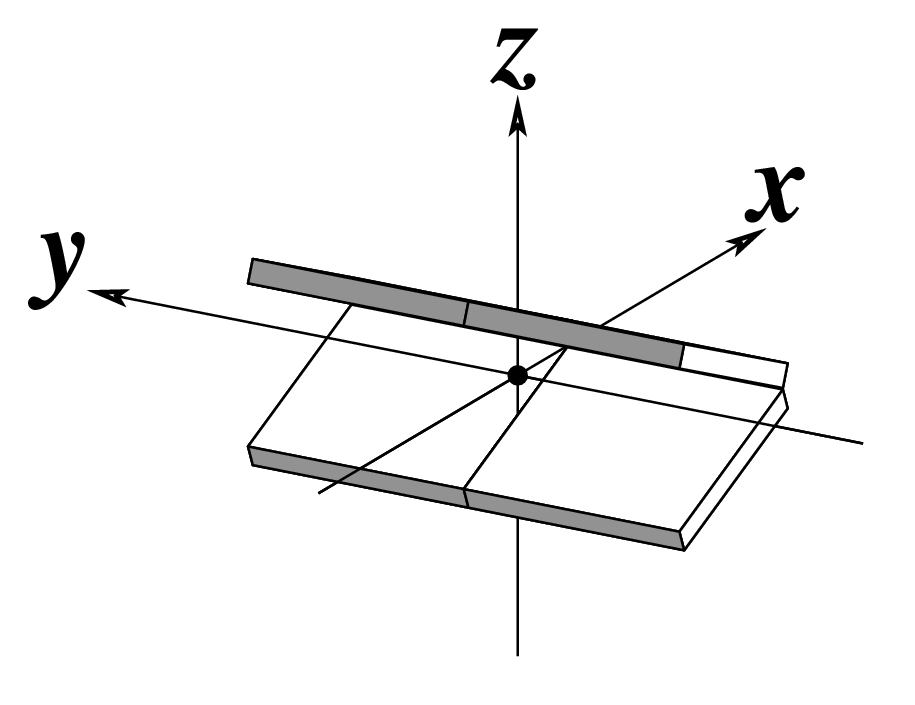 |
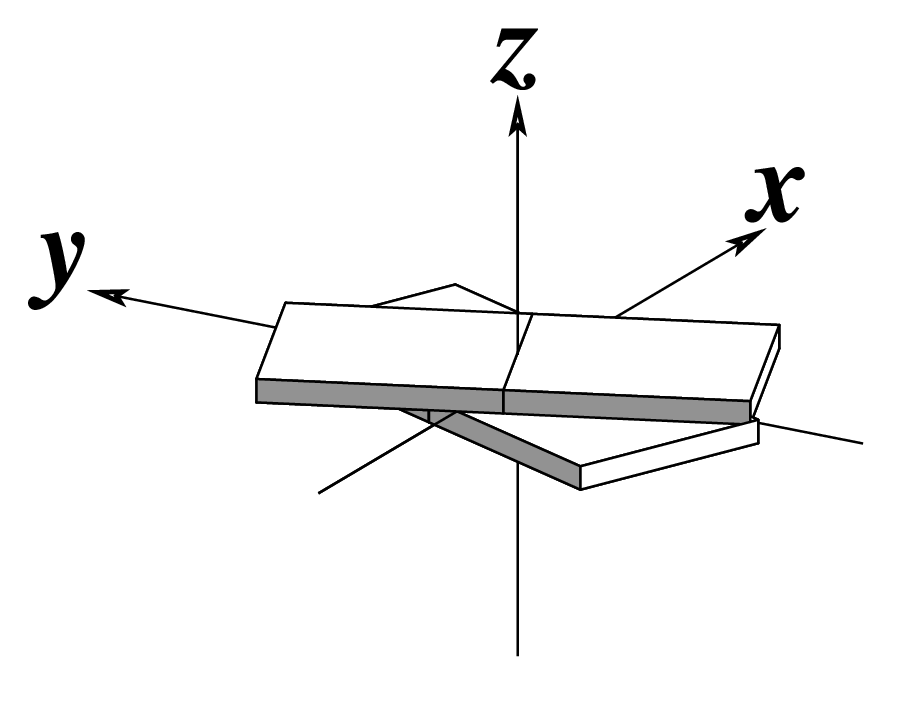 |
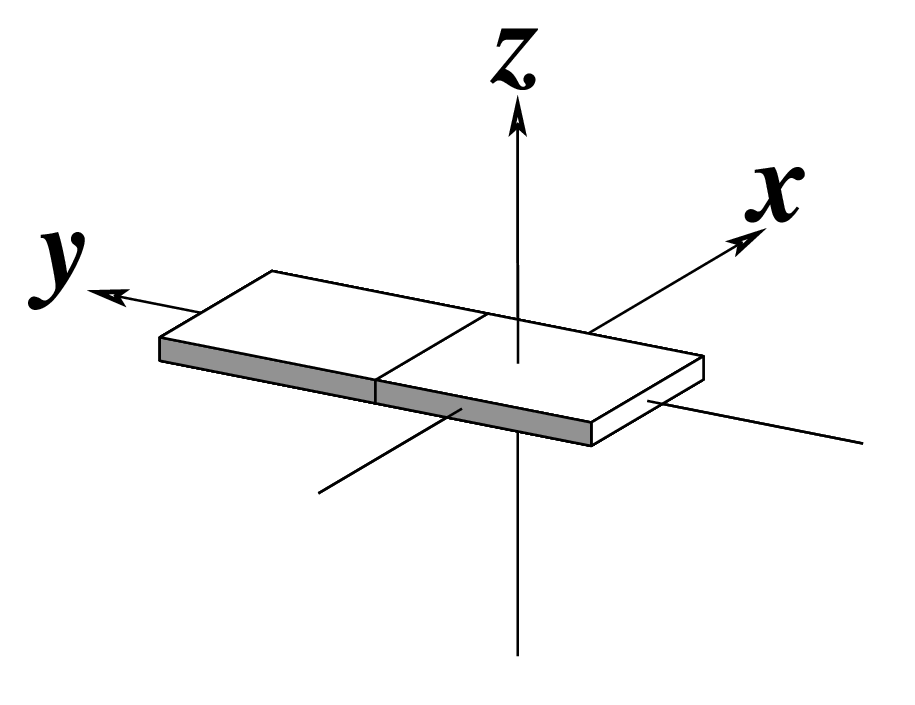
| x-displacement (dx) |
y-displacement (dy) |
Helical Rise (h) |
 |
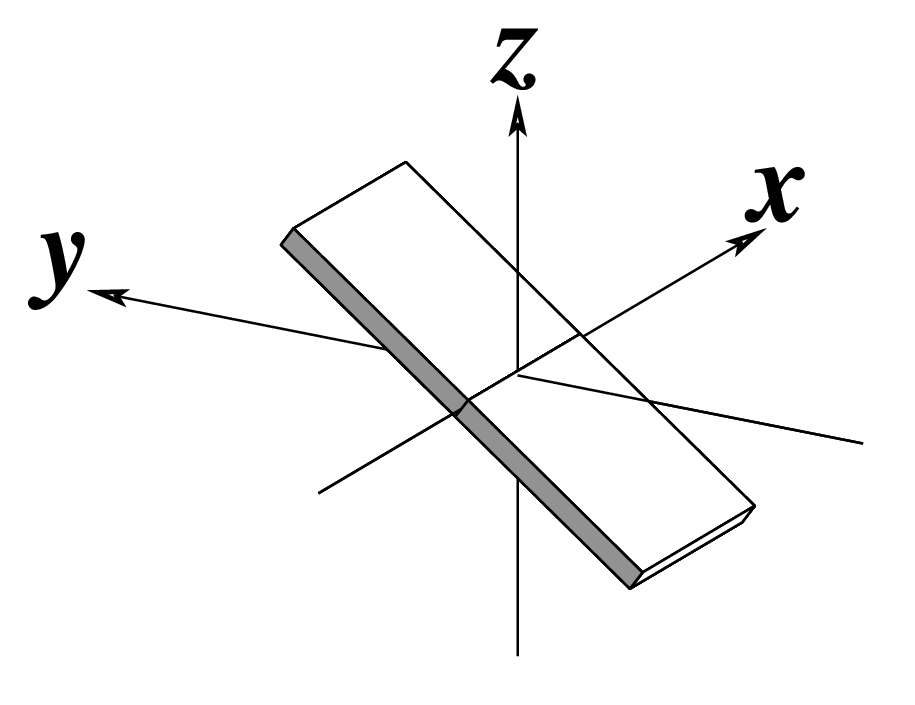
 |
As for Rise above
(for illustration purpose) |
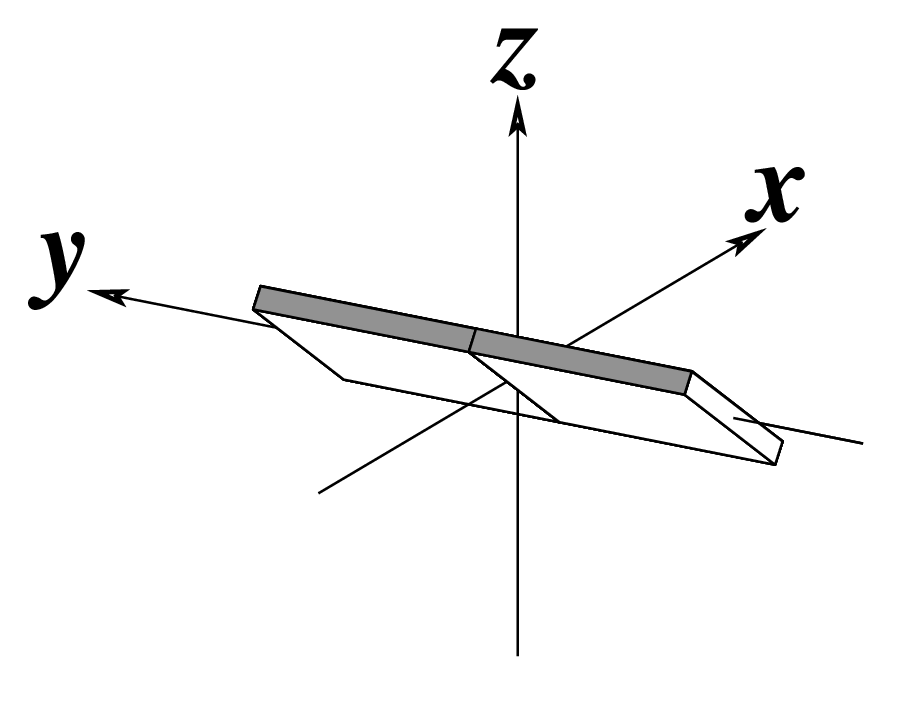
| Inclination (η) |
Tip (θ) |
Helical Twist (Ωh) |
 |
 |
As for Twist above
(for illustration purpose) |

Dear sir/Mam,
I am Supriyo Naskar, a Ph.D. student of the Indian Institute of Science, Bangalore India. For One of my research work, I need to calculate the helicoidal parameters of RNA. I would like to attach the schematic diagrams of helicoidal parameters as given in your website. I will cite the papers mentioned in the websites. Kindly let me know, whether I am allowed to do that?
Thanks in advance,
Best Regards,
Supriyo Naskar
— Supriyo Naskar ·
2019-04-15 15:48 ·
#
Yes, you’re welcome to use these schematic diagrams as long as the papers are properly cited.
Thanks,
Xiang-Jun
— Xiang-Jun Lu ·
2019-04-15 16:36 ·
#
Dear Dr. Lu,
I would like to reuse your figures above for a textbook that I am currently writing. I would appreciate your permission to reproduce. It will be properly cited. Regards,
Siddhartha Roy
— Prof. Siddhartha Roy ·
2020-02-22 02:55 ·
#
Dear Prof. Roy,
I’m glad that you’ve found the schematics useful to the textbook you’re writing. Please go-head using them as you see fit, and cite the 3DNA paper(s).
Please let’s know when your book is published. Viewers of the website (and 3DNA users in general) may well find it helpful.
Best regards,
— Xiang-Jun ·
2020-02-22 08:59 ·
#
hello sir,
I am a phd student. I want to use your web server to predict DNA 3D structure. so how do I get parameters for structure prediction.
thank you
— Abhigna ·
2020-03-01 23:32 ·
#
Please visit 3DNA Web 2.0 (http://web.x3dna.org) for ways to generating 3D nucleic-acid structures.
Thanks,
— Xiang-Jun ·
2020-04-18 21:00 ·
#
Hi,
How can I use the parameter file generated in your webpage in Phenix real space refinement. Is there a way to generate the restraints from your program. Your parameter file saves as .par which is not recognized by the Phenix. Any suggestions? thanks a lot
regards
Manoj Saxena
— MANOJ Saxena ·
2020-09-16 08:52 ·
#
Hi,
Thanks for stopping by and making a comment.
I am not sure how Phenix real space refinement works. Thus, I cannot offer any concrete suggestion on applying 3DNA parameters to Phenix. As you noticed, the 3DNA .par file is not directly applicable to generate the restraints for Phenix refinement.
You may ask Phenix developers and play around Phenix to know how the real space refinement works.
Best regards,
Xiang-Jun
— Xiang-Jun ·
2020-09-16 10:45 ·
#
« Reverse Watson-Crick base pairs
·
Classification of dinucleotide steps into A- and B- and TA-DNA »
Thank you for printing this article from http://x3dna.org/. Please
do not forget to visit back for more 3DNA-related information. —
Xiang-Jun Lu

















