I recently came across the Direk & Doluca (2024) paper on CIIS‐GQ: Computational Identification and Illustrative Standard for representation of unimolecular G‐Quadruplex secondary structures. Since DSSR is mentioned extensively in this work, with a section comparing CIIS-GQ and DSSR in supplementary materials, it is worthwhile to explore the issues raised in the paper. Overall, following literature allows me to clarify misconceptions and fix bugs that further improve DSSR.
The data which contain the G-quadruplexes were identified by DSSR-G4DB website [12, 13, 16]. All of the PDB (protein data bank) ids of DNA and RNA structures are extracted from the aforementioned website and the pdb files which contain the three dimensional data of the corresponding structures were downloaded from the protein data bank [3–5]. [under section "Materials and Methods": "Data"]
The DNA and RNA structures listed in 3DNA website were identified and downloaded from Protein Data Bank. Only unimolecular structures were used for the rest of the study (Supplementary Fig. 2). [under section "Results"]
Additionally, DSSR requires licensing to get annotation results for G-quadruplex structures. Fortunately, the annotation results for a number of G-quadruplexes were already published at DSSR-G4DB (46) and we were able to compare. [under section "Comparison with DSSR" in supplemental materials]
I am glad the DSSR-G4DB website served as a starting point for this study. The G4.x3dna.org website, where DSSR-G4DB is hosted, has always been available to the public. With the NIH R24GM153869 grant support, the standalone DSSR software is free for academic use and can be obtained from the Columbia Technology Ventures (CTV) website.
All obtained results for each pdb file were compared with DSSR. Out of which 35 DNA and 13 RNA structures were analyzed differently (Supplementary Table 2). Significant differences were detected for a number of structures between CIIS-GQ and DSSR analysis. For example, in 1k8p, 3ibk, 6ip7 and 5ccw structures, DSSR fails to identify some loops in some structures.
Most common issue that we have observed with DSSR is that it places loops in wrong places in some structures. For example, In 2a5p structure, the first loop is identified as reversal by both tools but DSSR also assigns the G6 to this loop which already participates in a tetrad. Such misplacement of tetrad-forming guanines in a loop is also seen in other structures such as 2a5r, 2kpr, 2m53, 2m92, etc (Supplementary Table 2).
The G4 module in DSSR was first developed around 2017-2018 and the work was mentioned briefly in the Lu (2020) paper on DSSR-PyMOL integration. However, due to the funding gap, the development of the G4 module was put on hold. I have never got a chance to write a paper documenting the detailed algorithms for the identification, annotation, and visualization of G-quadruplexes. I recently revamped the G4.x3dna.org website from inside out, and reprocessed all PDB structures to compile the DSSR-G4DB database. Along the way, the G4 module has been updated and improved. Now I'm actively working on a manuscript on the G4 module in DSSR and the associate website.
DSSR has clear definitions of G4-helix and G4-stem, and the corresponding loops. Specifically, for PDB entry 2a5p, DSSR reports the following:
## List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity,
and may contain more than one G4-stem.
##### Helix#1, 3 G-tetrads, INTRA-molecular, with 1 stem
1 glyco-bond=---- sugar=---- groove=---- WC-->Major O+ nts=4 GGGG A.DG4,A.DG8,A.DG13,A.DG17
2 glyco-bond=---- sugar=.--- groove=---- WC-->Major O+ nts=4 GGGG A.DG5,A.DG9,A.DG14,A.DG18
3 glyco-bond=-s-- sugar=-3-- groove=wn-- WC-->Major Z- nts=4 GGGG A.DG6,A.DG24,A.DG15,A.DG19
step#1 pm(>>,forward) area=9.64 rise=3.19 twist=32.7
step#2 pm(>>,forward) area=12.93 rise=3.29 twist=29.4
strand#1 DNA glyco-bond=--- sugar=-.- nts=3 GGG A.DG4,A.DG5,A.DG6
strand#2 DNA glyco-bond=--s sugar=--3 nts=3 GGG A.DG8,A.DG9,A.DG24
strand#3 DNA glyco-bond=--- sugar=--- nts=3 GGG A.DG13,A.DG14,A.DG15
strand#4 DNA glyco-bond=--- sugar=--- nts=3 GGG A.DG17,A.DG18,A.DG19Notice the differences in grooves between the first two G-tetrads vs the 3rd one, and the breaking backbone for strand#2 between G9 and G24.
## List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity.
Bulges are also allowed along each of the four strands.
##### Stem#1, 2 G-tetrads, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
1 glyco-bond=---- sugar=---- groove=---- WC-->Major O+ nts=4 GGGG A.DG4,A.DG8,A.DG13,A.DG17
2 glyco-bond=---- sugar=.--- groove=---- WC-->Major O+ nts=4 GGGG A.DG5,A.DG9,A.DG14,A.DG18
step#1 pm(>>,forward) area=9.64 rise=3.19 twist=32.7
strand#1 U DNA glyco-bond=-- sugar=-. nts=2 GG A.DG4,A.DG5
strand#2 U DNA glyco-bond=-- sugar=-- nts=2 GG A.DG8,A.DG9
strand#3 U DNA glyco-bond=-- sugar=-- nts=2 GG A.DG13,A.DG14
strand#4 U DNA glyco-bond=-- sugar=-- nts=2 GG A.DG17,A.DG18
loop#1 type=propeller strands=[#1,#2] nts=2 GT A.DG6,A.DT7
loop#2 type=propeller strands=[#2,#3] nts=3 gGA A.DI10,A.DG11,A.DA12
loop#3 type=propeller strands=[#3,#4] nts=2 GT A.DG15,A.DT16Thus the G4-stem consists of two G-tetrads only, and G6 which is part of the 3rd G-tetrad becomes part of a propeller loop. Similar arrangement applies to the other cases.
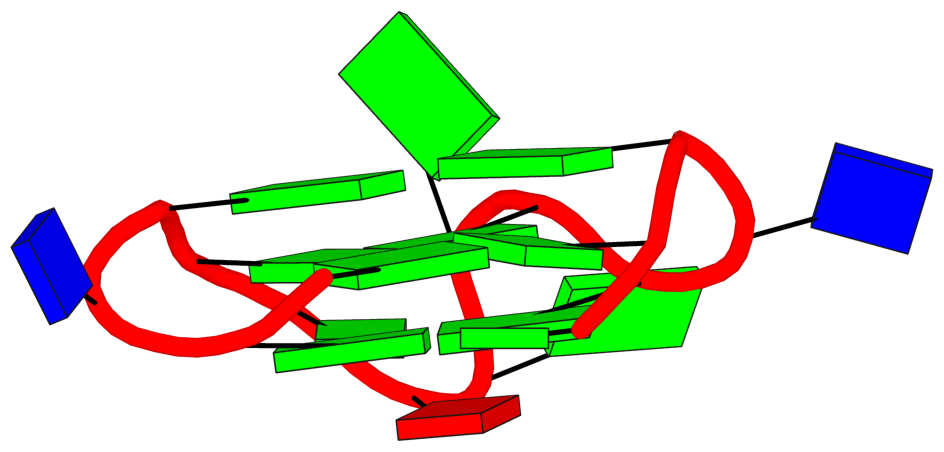 DSSR also reports the following loop:
DSSR also reports the following loop:
## List of 1 non-stem G4-loop (including the two closing Gs)
1 type=diagonal helix=#1 nts=6 GGAAGG A.DG19,A.DG20,A.DA21,A.DA22,A.DG23,A.DG24In my understanding, the definition and nomenclature of loops in G4 structures are not yet standardized. I am monitoring the development in this field and will update DSSR as needed in due course.
There may also be different types of loops identified by these tools. For example, in 1oz8, which is depicted by CIISGQ as two separate G4s, DSSR fails to identify the G-tetrad, [2, 5, 8, 11], that lies on the outside of the structure. This results in identification of loops formed within this tetrad and its stacking neighbor different to CIIS-GQ. While CIISGQ identifies these loops as reversal, just like the other loops in the structure, DSSR identifies them as non-stem lateral loops. This causes complete misinterpretation of the size and the type of loops in the structure.
The revised DSSR output for PDB entry 1oz8 has the G-tetrad A.DG2,A.DG5,A.DG8,A.DG11 manually added as part of the input, and now all three propeller loops are correctly identified. By default, G11 does not form proper G+G pairs (of LW type cWH or cHW, and Saenger type VI) with G2 and G8. The distortion of the G-tetrad is obvious in the block representation of the structure.
In 4u5m, similar to 1oz8, the structure may be interpreted as two separate G4s connected through a single link (T13,T14). In this case, DSSR identifies only two loops in one of the G4s and labels them as non-stem V-shaped loops. This also differs from CIIS-GQ where CIIS-GQ interprets all loops in both G4 as reversal. Structures containing multiple G4s, such as 1oz8, 4u5m and 6kvb, are often identified with different loop types by DSSR, while CIIS-GQ can recognise the loops correctly and simplifies the comprehension of the structure.
For PDB entry 4u5m, the same arguments above regarding the G4-stem and loops for 2a5p apply.
1 glyco-bond=s--- sugar=---- groove=w--n Major-->WC O+ nts=4 GGGG A.DG2,A.DG11,A.DG8,A.DG5
2 glyco-bond=---- sugar=---- groove=---- Major-->WC O+ nts=4 GGGG A.DG3,A.DG12,A.DG9,A.DG6
3 glyco-bond=---- sugar=---- groove=---- WC-->Major O+ nts=4 GGGG A.DG24,A.DG15,A.DG18,A.DG21
4 glyco-bond=---- sugar=---- groove=---- WC-->Major O+ nts=4 GGGG A.DG23,A.DG26,A.DG17,A.DG20As shown, the backbone between G15 and G26 is broken. Moreover, here the assignment of Gs along the strand may need to be manually adjusted.
As shown in Table 1, by relaxing angle and distance parameters, we were able to identify more tetrads (6T2G, 1OZ8) than DSSR, which detects them as multiplets instead.
The current DSSR results for PDB entry 6t2g and 1oz8 are all as expected. Moreover, DSSR can handle PDB entry 6t2g automatically, while for PDB entry 1oz8 user needs to manually edit the input to include the G-tetrad with G11. By allowing users to specify tetrads, DSSR offers precise control and great flexibility, e.g., to include the G-C-G-C tetrads in PDB entry 1a6h.
DSSR has a detailed explanation of strands, tetrads and loops. However, the comprehensive output of DSSR is often hard to understand and grasp the details of the structure. [in supplemental materials]
The detailed explanations are provided to help users understand the DSSR output. They are most insightful in combination with the schematic block diagrams. For examples, for PDB entry 1a6h, the middle G-C-G-C tetrads are crystal clear with the long green and yellow rectangular blocks, specially along with the detailed annotations of the tetrads, as shown below.
1 glyco-bond=s-s- sugar=---. groove=wnwn Major-->WC -- nts=4 GGGG A.DG1,A.DG11,B.DG8,B.DG4
2 glyco-bond=---- sugar=-.-- groove=---- -- -- nts=4 CGCG A.DC2,A.DG10,B.DC9,B.DG3
3 glyco-bond=---- sugar=--.- groove=---- -- -- nts=4 GCGC A.DG3,A.DC9,B.DG10,B.DC2
4 glyco-bond=-s-s sugar=---- groove=wnwn WC-->Major -- nts=4 GGGG A.DG4,A.DG8,B.DG11,B.DG1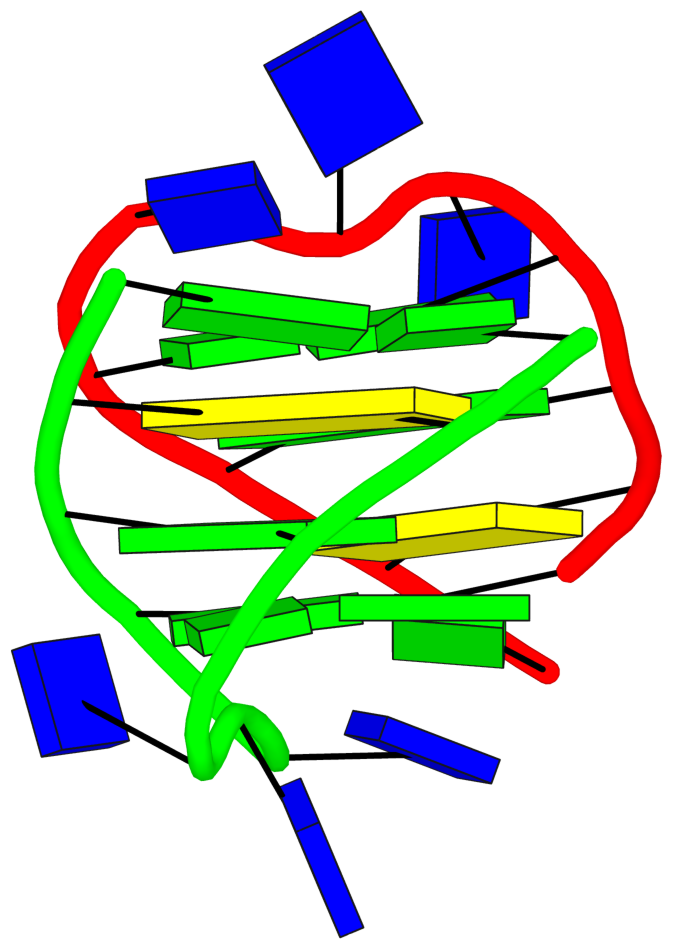
Another advantage of CIIS-GQ is that it requires only two thresholds, the thresholds of distance and angle parameters that can be modified to detect loosely connected tetrads. Due to this advantage, the identification of the tetrads were possible in at least two structures. In case of 1OZ8, DSSR found three tetrads (G1-G4-G7-G10, G13-G16-G19-G22 and G14-G17-G20-G23) as shown at the result page2 (47) while CIIS-GQ has found one more tetrad which is G2-G5-G8-G11. In comparison DSSR highlighted G5-G8-G11 as a multiplet, omitting the G2. Based on this difference, loop classification differs with CIIS-GQ. DSSR has identified 3 stem reversal loops and 3 non-stem lateral loops while we have identified 7 reversal loops. Stem loop is defined as any loop that also forms a duplex within itself.
DSSR now has PDB entry 1oz8 properly characterized, by manually adding the G-tetrad involving G11, as detailed above.
A similar difference exists in 6T2G. DSSR could find 2 tetrads in this structure (G2-G6-G11-G26 and G4-G9-G13-G28) as shown at the result page3 (47) while CIIS-GQ found one more tetrad, G3-G7-G12-G27. DSSR is able to show these four guanines as a multiplet in the list of multiplets section, however does not present it as a tetrad like the other two tetrads. As a result, CIIS-GQ loop types and placements are also different. DSSR has found six lateral loops while CIIS-GQ has found three reversal loops.
DSSR can now handle PDB entry 6t2g automatically. Previous versions of DSSR missed the G-tetrad (G3+G7+G12+G27) because of the G12+G27 pair: it fails the criteria to be classified as the pair of LW type cWH or cHW and Saenger type VI. Thus G3+G7+G12+G27 do not qualify as a G-tetrad, but they still form a multiplet with four guanines.
References
Direk, T., & Doluca, O. (2024). Computational Identification and Illustrative Standard for Representation of Unimolecular G-Quadruplex Secondary Structures (CIIS-GQ). Journal of Computer-Aided Molecular Design, 38(1), 35. https://doi.org/10.1007/s10822-024-00573-1
Lu, X.-J. (2020). DSSR-enabled innovative schematics of 3D nucleic acid structures with PyMOL. Nucleic Acids Research, gkaa426. https://doi.org/10.1093/nar/gkaa426
