Any analysis of nucleic acid structures start with the identification of nucleotides (nts), the basic building unit. As per the PDB convention, each nt (like any other ligands) is specified by a three-letter identifier. For example, the four standard RNA nts are ..A, ..C, ..G, and ..U, respectively. The four corresponding standard DNA nts are .DA, .DC, .DG, and .DT, respectively. Note that here, for visualization purpose, each space is represented by a dot (.). In practice, the following codes for the five standard DNA/RNA nts — ADE, CYT, GUA, THY, and URA — are also commonly encountered, among other variants.
On top of the standard nts, there are numerous modified ones, each assigned a unique three-letter code. In the classic yeast phenylalanine tRNA (PDB id: 1ehz), 14 out of the 76 nts are modified, as shown in Fig. 1 below.
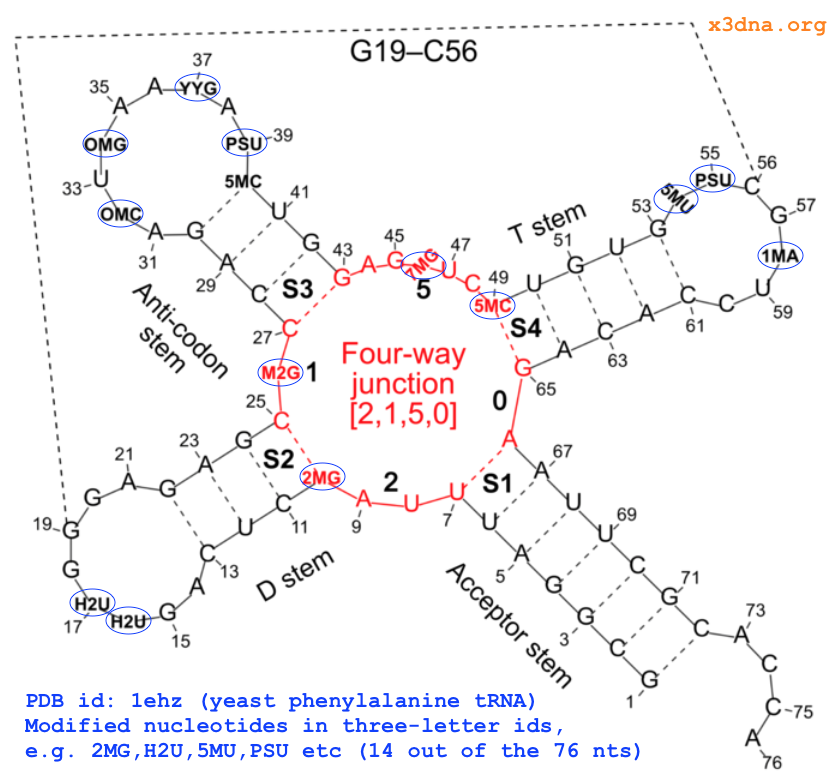
Fig. 1: Modified nucleotides in yeast phenylalanine tRNA 1ehz
It is challenging to maintain a comprehensive and updated list of ever-inceasing nts encountered in the PDB and molecular dynamics (MD) simulation packages (e.g., AMBER, GROMACS, and CHARMM). Thus, as of today, some well-known DNA/RNA structural bioinformatics tools can handle only standard nts or a limited list of modified ones.
From early on in the development of 3DNA, I observed that all recognized nts have a core six-membered ring, with atoms named N1,C2,N3,C4,C5,C6 consecutively (see Fig. 2 below). Purines have three additional atoms, named N7,C8,N9. So it is feasible to automatically identify nts, and classify them as pyrimidines and purines, based on the common core skeleton shared by all of them. Moreover, the ‘skeleton’ is not effected by any possible tautomeric or protonation state.
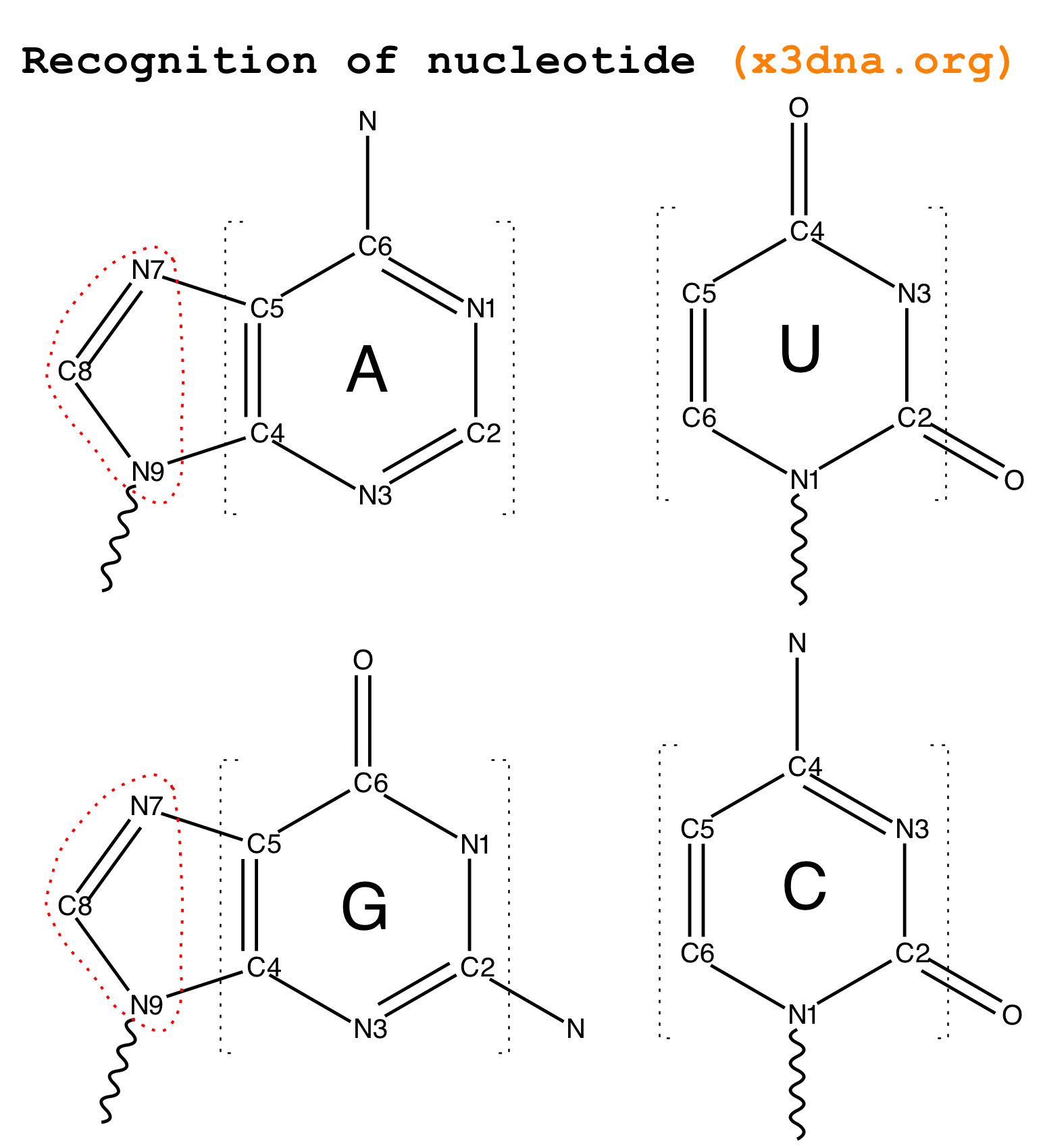
Fig. 2: Identification of nts in 3DNA/DSSR based on atomic names and planar geometry
Early versions of 3DNA employed only three atoms (N1, C2 and C6) and three distances to decide a nt. Purines were further discriminated by the N9 atom, and the N1–N9 distance. While developing DSSR, I revised the nt-identification algorithm by using a least-squares fitting procedure that makes use of all available base ring atoms instead of selected ones. The same new algorithm has also been adapted into the find_pair/analyze etc programs in 3DNA, as of v2.2.
As always, the idea can be best illustrated with a worked example. Guanine in its standard base reference frame, with the following list of nine ring atoms coordinates, is chosen for the least-squares fitting. See file Atomic_G.pdb in the 3DNA distribution, and also Table 1 of the report A Standard Reference Frame for the Description of Nucleic Acid Base-pair Geometry.
ATOM 2 N9 G A 1 -1.289 4.551 0.000 ATOM 3 C8 G A 1 0.023 4.962 0.000 ATOM 4 N7 G A 1 0.870 3.969 0.000 ATOM 5 C5 G A 1 0.071 2.833 0.000 ATOM 6 C6 G A 1 0.424 1.460 0.000 ATOM 8 N1 G A 1 -0.700 0.641 0.000 ATOM 9 C2 G A 1 -1.999 1.087 0.000 ATOM 11 N3 G A 1 -2.342 2.364 0.001 ATOM 12 C4 G A 1 -1.265 3.177 0.000
By using a ls-fitting procedure, only (any) three atoms are needed. We no longer need to make explicit selection, as we did previously (N1,C2,C6 and N9), thus allowing for possible modification on these atoms.
Using four nts (G1, 2MG10, H2U16, and PSU39, see Fig. 1 above top) of 1ehz as examples, the following list gives the atomic coordinates of base ring atoms, and root-mean-squres devisions (rmsd) of the least-squares fit. Of course, when performing least-squares fitting, the names of corresponding atoms must match (note the different ordering of atoms for H2U and PSU in the list vs the above standard G reference).
#G1, rmsd=0.008 ATOM 14 N9 G A 1 51.628 45.992 53.798 1.00 93.67 N ATOM 15 C8 G A 1 51.064 46.007 52.547 1.00 92.60 C ATOM 16 N7 G A 1 51.379 44.966 51.831 1.00 91.19 N ATOM 17 C5 G A 1 52.197 44.218 52.658 1.00 91.47 C ATOM 18 C6 G A 1 52.848 42.992 52.425 1.00 90.68 C ATOM 20 N1 G A 1 53.588 42.588 53.534 1.00 90.71 N ATOM 21 C2 G A 1 53.685 43.282 54.716 1.00 91.21 C ATOM 23 N3 G A 1 53.077 44.429 54.946 1.00 91.92 N ATOM 24 C4 G A 1 52.356 44.836 53.879 1.00 92.62 C
#2MG10, rmsd=0.018 HETATM 207 N9 2MG A 10 61.581 47.402 18.752 1.00 42.14 N HETATM 208 C8 2MG A 10 62.199 48.621 18.635 1.00 40.38 C HETATM 209 N7 2MG A 10 63.494 48.534 18.422 1.00 40.70 N HETATM 210 C5 2MG A 10 63.745 47.167 18.395 1.00 43.82 C HETATM 211 C6 2MG A 10 64.965 46.449 18.205 1.00 43.45 C HETATM 213 N1 2MG A 10 64.767 45.086 18.293 1.00 44.71 N HETATM 214 C2 2MG A 10 63.541 44.482 18.486 1.00 47.21 C HETATM 217 N3 2MG A 10 62.411 45.125 18.614 1.00 45.85 N HETATM 218 C4 2MG A 10 62.574 46.451 18.582 1.00 43.27 C
#H2U16, rmsd=0.188 HETATM 336 N1 H2U A 16 77.347 53.323 34.582 1.00 91.19 N HETATM 337 C2 H2U A 16 76.119 52.865 34.160 1.00 92.39 C HETATM 339 N3 H2U A 16 75.123 52.894 35.107 1.00 93.28 N HETATM 340 C4 H2U A 16 75.289 52.711 36.458 1.00 93.34 C HETATM 342 C5 H2U A 16 76.696 52.479 36.909 1.00 93.77 C HETATM 343 C6 H2U A 16 77.717 53.238 36.039 1.00 93.22 C
#PSU39, rmsd=0.004 HETATM 845 N1 PSU A 39 74.080 36.066 5.459 1.00 75.82 N HETATM 846 C2 PSU A 39 74.415 36.835 4.354 1.00 75.59 C HETATM 847 N3 PSU A 39 75.735 36.769 3.984 1.00 76.29 N HETATM 848 C4 PSU A 39 76.728 36.038 4.591 1.00 77.28 C HETATM 849 C5 PSU A 39 76.307 35.280 5.732 1.00 77.93 C HETATM 850 C6 PSU A 39 75.025 35.316 6.112 1.00 76.07 C
As noted in the DSSR paper, the rmsd is normally <0.1 Å since base rings are rigid. To account for experimental error and special non-planar cases, such as H2U in 1ehz, the default rmsd cutoff is set to 0.28 Å by default.
With the above detailed algorithm, DSSR (and the 3DNA find_pair/analyze programs) can automatically identify virtually all ‘recognizable’ nts in the PDB. A survey performed in June 2015 detected 630 different types of modified nucleotides in the PDB.
It is worth noting the following points:
- The choice of standard G instead of A as the reference base has no impact on the results. As a matter of fact, the rmsd between G and A is only 0.04 Å. Note also the generous default cutoff of 0.28 Å.
- The method obviously depends on proper naming of the ring atoms. Specially, the base ring atoms must be named
N1,C2,N3,C4,C5,C6consecutively, with purines having three additional atoms namedN7,C8,N9. Thus, under this scheme, TPP (thiamine diphosphate) would not be recognized as a nt by default, simply because of the extra prime (′) of atoms in the six-membered ring. In nucleic acid structures, the prime symbol is normally associated with atoms of the sugar moiety (e.g., the C5′ atom).
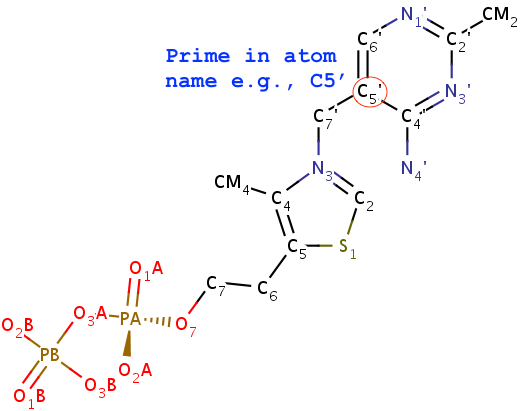
Fig. 3: TPP (thiamine diphosphate) would not be recognized as a nt.
- On the other hand, nt cofactors in an otherwise ‘pure’ protein structure will also be recognized. One example is the two AMP (adenosine monophosphate) ligands in PDB entry 12as. This extra identification of nts does no harm in such cases. As shown in the analysis of the SAM-I riboswitch in the DSSR paper, taking the SAM ligand as a nt in base triplet recognition is a neat feature.
- Once a nucleotide has been identified and classified into purines and pyrimidines, exocyclic atoms can be used for further assignment:
O6orN2distinguishes guanine from adenine,N4separates cytosine from thymine and uracil, andC7(orC5M, the methyl group) differentiates thymine from uracil. For some modified nts, the distinctions within purines or pyrimidines may not be that obvious. For example, inosine may be taken as a modified guanine or adenine. However, this ambiguity does not pose any significant effect on the calculated base-pair parameters.
- In DSSR and 3DNA, each identified nt is assigned a one-letter shorthand code: the standard
..A,.DA, andADE(among a few other common variations) is shortened to upper-caseA, and similarly forC,G,T, andU. Modified nts, on the other hand, are shortened to their corresponding lower-case symbol. For example, modified guanine such as2MGandM2Gin the yeast phenylalanine tRNA (see Fig. 1 above) is assignedg. So in 3DNA/DSSR output, the upper and lower cases of bases (e.g.,nts=3 gCG A.2MG10,A.C25,A.G45) convey special meanings.
