This post is a recap of the recently introduced ‘simple’ base-pair (bp) parameters (Fig. 1) useful for describing non-Waton-Crick pairs, and the highly effective cartoon-block representations of nucleic acid structures. Both features are readily available from 3DNA/DSSR, as detailed here using four examples of representative DNA/RNA structures (Fig. 2). Links to related blog posts are provided at the end.
Note added on Feb. 2, 2016: in fact, this post had been intended to supplement a short communication titled Characterization of base-pair geometry that Dr. Wilma Olson and I recently contributed to the January 2016 issue of Computational Crystallography Newsletter (CCN). That’s why the URL of this post is ‘http://x3dna.org/highlights/CCN-on-base-pair-geometry’ instead of what one would expect from the title. The data files, scripts, images, and linked herein should enable interested users a thorough understanding of the ‘simple’ base-pair parameters. If you have problems in reproducing our reported results, please do not hesitate to let me know (publicly). You are welcome to either leave comments to this post or ask any related questions on the 3DNA Forum.
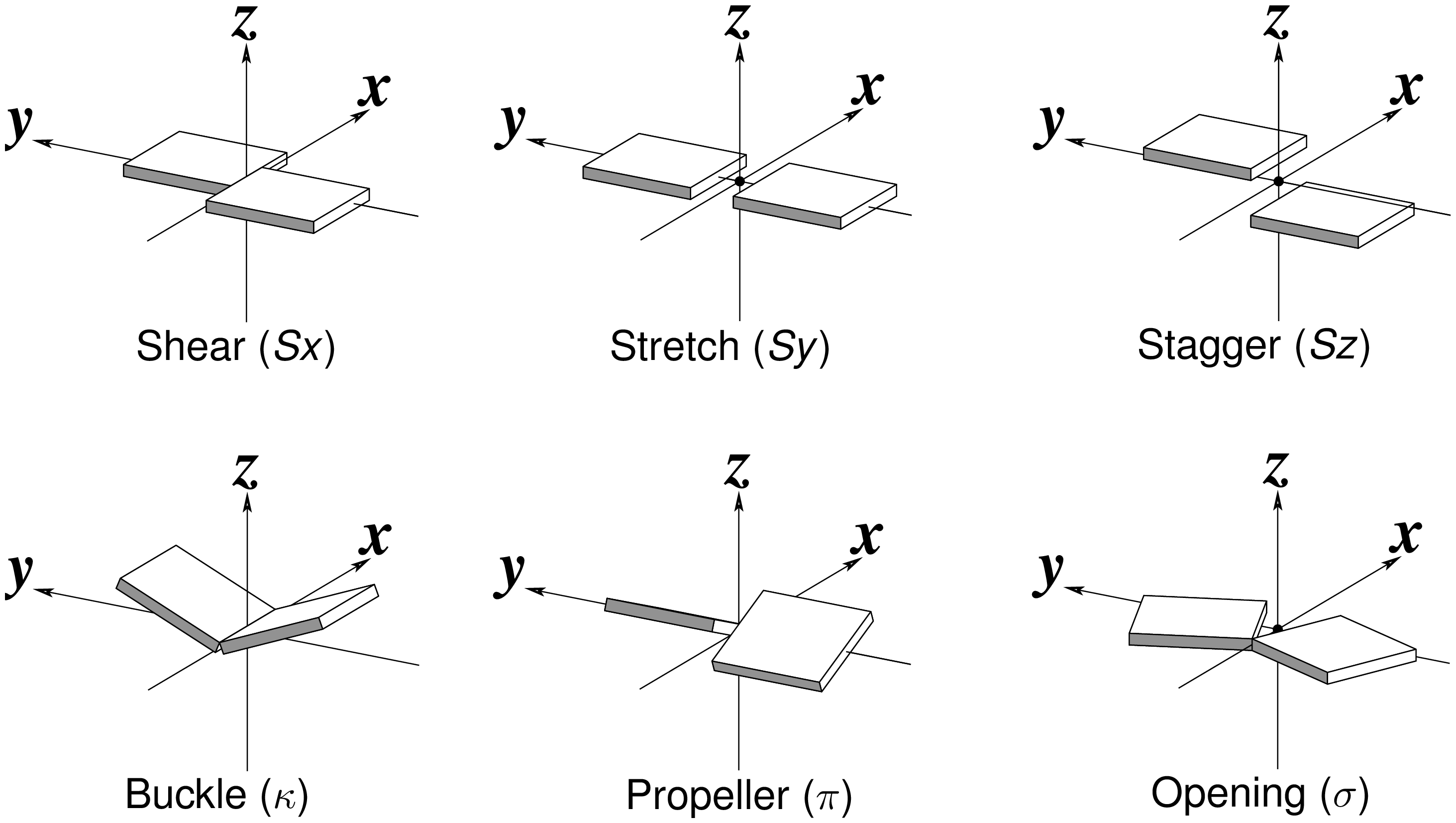
Six rigid-body parameters
Fig. 1: Schematic diagrams of the six rigid-body parameters commonly used for the characterization of base-pair geometry.
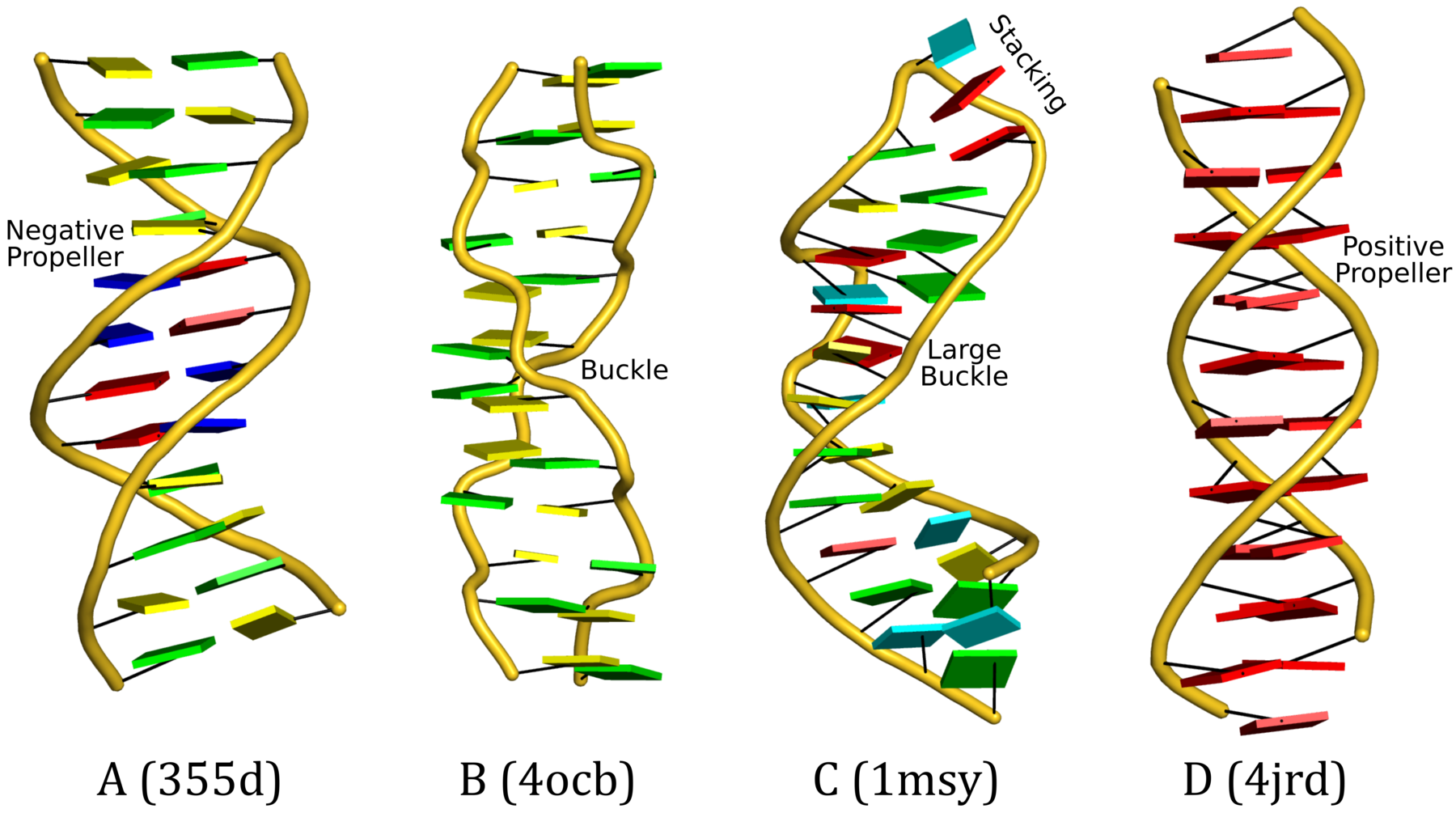
Cartoon-block representations
Fig. 2: DSSR-introduced cartoon-block representations of DNA and RNA structures that combine PyMOL cartoon schematics with color-coded rectangular base blocks: A, red; C, yellow; G, green; T, blue; and U, cyan. (A) The Dickerson B-DNA dodecamer solved at 1.4-Å resolution [PDB id: 355d (Shui et al., 1998)], with significant negative Propeller. (B) The Z-DNA dodecamer [PDB id: 4ocb (Luo et al., 2014)], with virtually co-planar C–G pairs at the ends, and noticeable Buckle in the middle. © The GUAA tetraloop mutant of the sarcin/ricin domain from E. coli 23 S rRNA [PDB id: 1msy (Correll et al., 2003)], with large Buckle in the A+C pair, and base-stacking interactions of UAA in the GUAA tetraloop (upper-right corner). (D) The parallel double-stranded poly(A) RNA helix [PDB id: 4jrd (Safaee et al., 2013)], with up to +14° Propeller. The simple, informative cartoon-block representations facilitate understanding of the base interactions in small to mid-sized nucleic acid structures like these. The base identity, pairing geometry, and stacking interactions are obvious.
Scripts and data files (Lu-CCN-examples.tar.gz)
find_pair 355d.pdb | analyze # 355d.out x3dna-dssr -i=355d.pdb -more -o=355d-dssr.out x3dna-dssr -i=355d.pdb --cartoon-block -o=355d.pml find_pair 4jrd.pdb | analyze # 4jrd.out x3dna-dssr -i=4jrd.pdb -more -o=4jrd-dssr.out x3dna-dssr -i=4jrd.pdb --cartoon-block -o=4jrd.pml find_pair 1msy.pdb | analyze # 1msy.out x3dna-dssr -i=1msy.pdb -more -o=355d-dssr.out x3dna-dssr -i=1msy.pdb --cartoon-block -o=1msy.pml find_pair --symm 4ocb.pdb1 | analyze --symm # 4ocb.out x3dna-dssr -i=4ocb.pdb1 --symm -more -o=4ocb-dssr.out x3dna-dssr -i=4ocb.pdb1 --symm --cartoon-block -o=4ocb.pml
Please note the following points:
- The above examples are based on 3DNA
v2.3-2016jan20and DSSRv1.4.8-2016jan16. - All data files (including PyMOL ray-traced PNG images used in Fig. 2) are packed into a tarball named Lu-CCN-examples.tar.gz for download.
- For PDB entry 4ocb, the biological unit (with suffix
.pdb1) is used to get a complete duplex structure. The symm option must be specified. - PDB files are used in the above illustration. In fact, the corresponding mmCIF files (
.cif) also work just fine. - The DSSR-derived .pml files can be fed into PyMOL for rendering. In addition to the directly generated
*.pmlfiles (e.g.,355d.pml), the PyMOL transformed version (i.e.,orient; turn z, -90) are also included, with names*-orient.pml(e.g.,355d-orient.pml). The PNG images (as shown in Fig. 2) are ray-traced using these reoriented pml files for the most extended vertical view. - The ‘simple’ base-pair parameters for 4jrd is shown below.
This structure contains 10 non-Watson-Crick (with leading *) base pair(s)
----------------------------------------------------------------------------
Simple base-pair parameters based on RC8--YC6 vectors
bp Shear Stretch Stagger Buckle Propeller Opening angle
* 1 A+A -7.96 0.41 -0.03 -13.64 -4.06 -179.47 14.2
* 2 A+A -7.86 0.38 -0.33 -10.20 -3.53 -179.34 10.8
* 3 A+A -7.96 0.43 0.02 -10.15 5.23 179.91 11.4
* 4 A+A -7.95 0.50 0.10 -9.24 8.04 179.15 12.2
* 5 A+A -7.95 0.46 0.08 -7.36 10.12 -179.98 12.5
* 6 A+A -7.97 0.60 0.06 -5.15 12.87 -176.75 13.9
* 7 A+A -7.88 0.66 -0.02 -7.82 11.89 -179.55 14.2
* 8 A+A -7.91 0.56 -0.05 -7.03 13.68 179.22 15.4
* 9 A+A -7.94 0.47 -0.03 -3.78 13.76 -179.24 14.3
* 10 A+A -7.92 0.42 0.10 -3.03 4.34 -178.91 5.3
Related posts
- Details on the simple base-pair parameters
- DSSR base blocks in PyMOL, interactively
- Fitting of base reference frame
- Automatic identification of nucleotides
- ‘Simple’ parameters for non-Watson-Crick base pairs
- Simple base-pair parameters
- Quantifying base-pair geometry by six rigid-body parameters
- How to calculate torsion angle?
- RNA cartoon-block representations with PyMOL and DSSR
- The DSSR User Manual (PDF)