In early 2015, Thomas Holder (the PyMOL Principal Developer at Schrodinger) and I agreed to work together on connecting DSSR to PyMOL. Moreover, we called for the community’s involvement in writing a DSSR plugin for PyMOL and received a few enthusiastic replies. Over the past few months, many significant progresses have been made in DSSR, including an article titled DSSR: an integrated software tool for dissecting the spatial structure of RNA published in Nucleic Acids Research (NAR) and a more streamlined DSSR-Jmol integration based on the --json output.
From the very beginning, Thomas and I had envisioned that the DSSR-PyMOL integration would include two components: one is to bring DSSR-derived RNA/DNA structural features into PyMOL (similar to the DSSR-Jmol interface, funcationality-wise), and the other is to render DSSR’s simple yet informative base-rectangular representations with PyMOL. While the ‘analysis’ component is a work in progress, the ‘visualization’ part is ready for the community to take advantage of.
Thomas has written a Python script named dssr_block.py. When the script is run in PyMOL, it adds the “dssr_block” command. The dssr_block.py script is less than 100 lines including documentation, with the real code taking no more than half of the total line number. The detailed documentation section (with two examples), when condensed, is as follows:
DESCRIPTION
Create a nucleid acid cartoon with DSSR
USAGE
dssr_block [selection [, state [, block_file [, block_depth [, name [, exe]]]]]]
ARGUMENTS
selection = str: atom selection {default: all}
state = int: object state (0 for all states) {default: -1, current state}
block_file = face|edge|wc|equal|minor|gray {default: face}
block_depth = float: thickness of rectangular blocks {default: 0.5}
name = str: name of new CGO object {default: dssr_block##}
exe = str: path to "x3dna-dssr" executable {default: x3dna-dssr}
EXAMPLE
fetch 1ehz, async=0
as cartoon
dssr_block
set cartoon_ladder_radius, 0.1
set cartoon_ladder_color, gray
set cartoon_nucleic_acid_mode, 1
# multi-state
fetch 2n2d, async=0
dssr_block 2n2d, 0
set all_states
Download the dssr_block.py script into a folder (directory) of your choice. Within PyMOL command window, type:
run dssr_block.py # to make the 'dssr_block' command avaible help dssr_block # to get the help message, with contents shown above
The resultant cartoon-block image for running the documented commands (except for the additional orient command for best view) for case 1ehz is shown in Fig. 1 below.
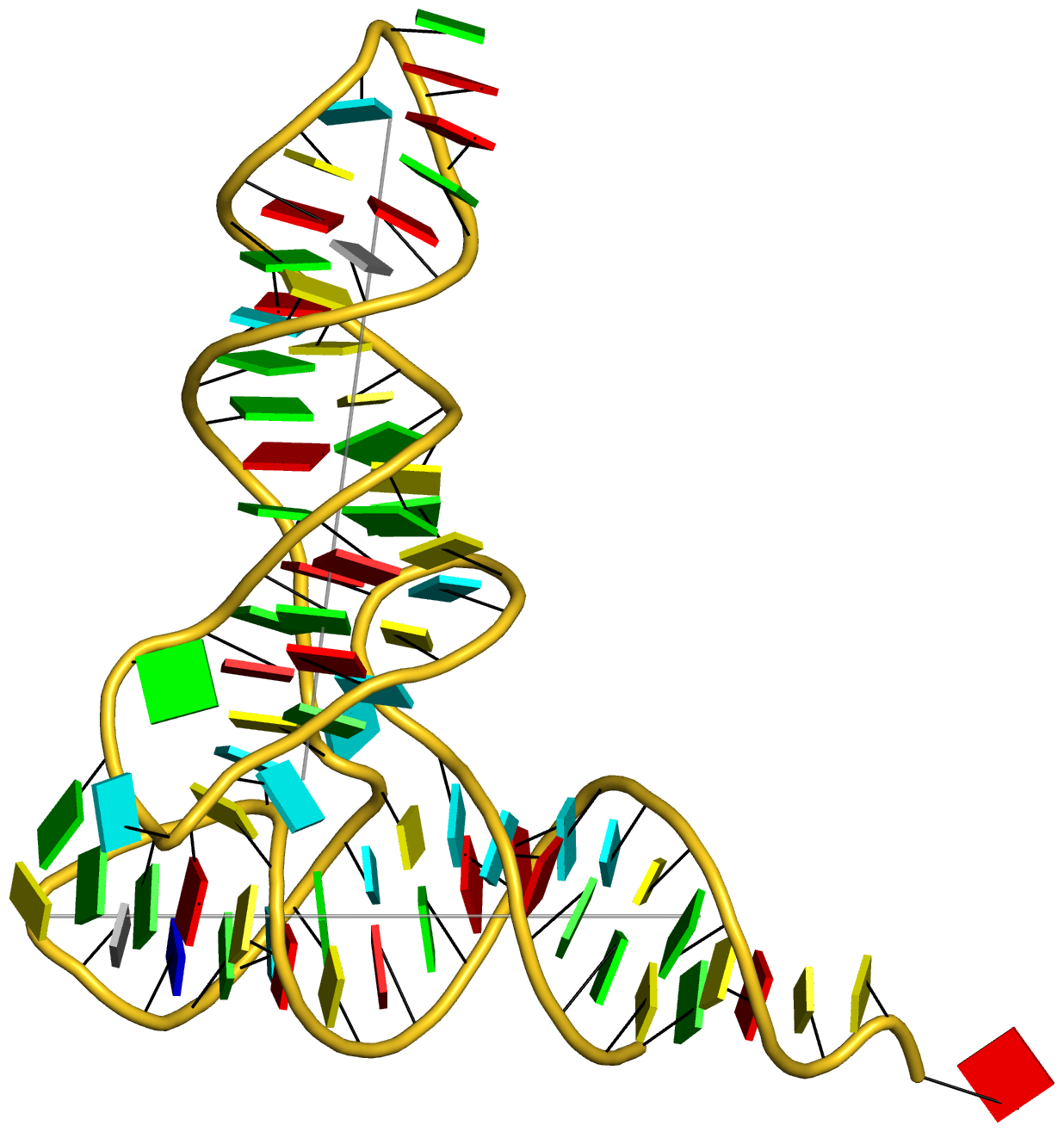
Fig. 1: Cartoon-block image generated by dssr_block.py for PDB entry 1ehz (yeast phenylalanine tRNA)
For the NMR ensemble 2n2d, the corresponding image (after running orient) is illustrated in Fig. 2 as follows:
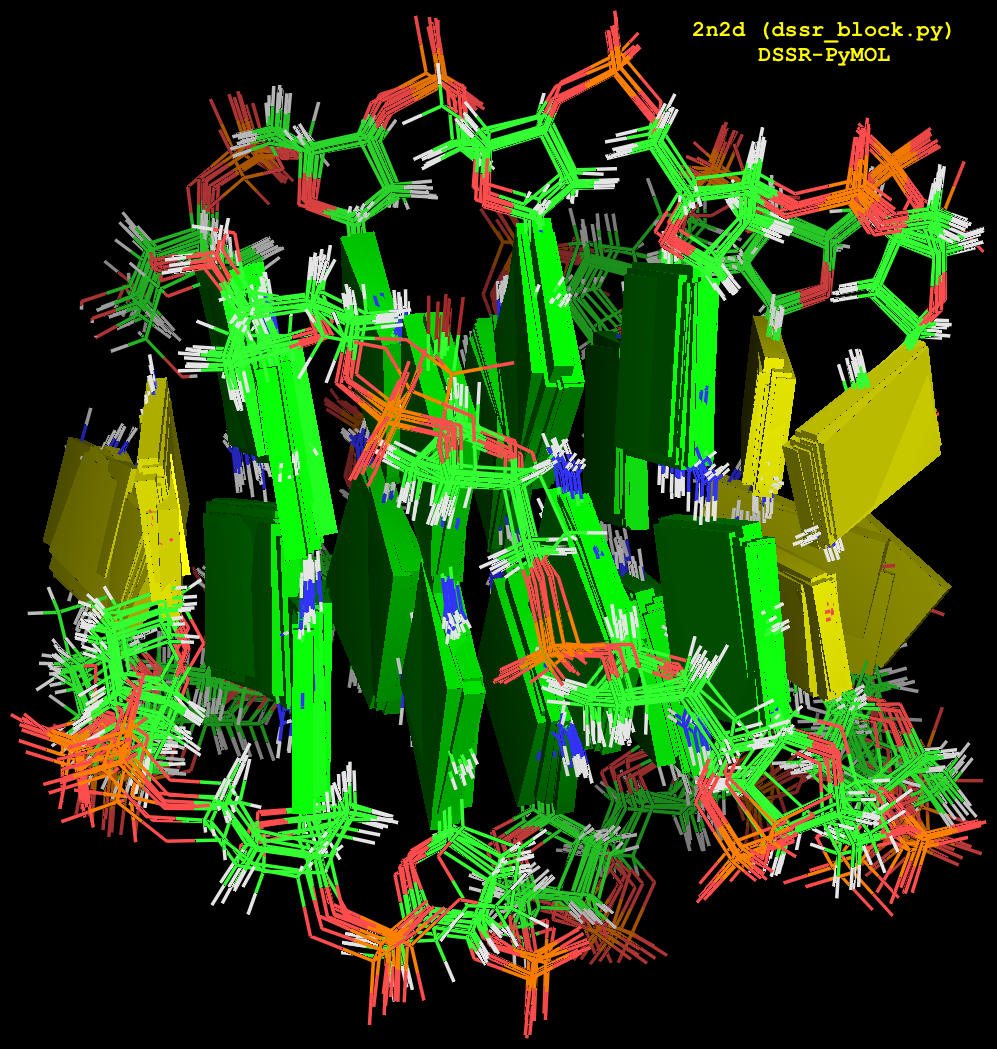
Fig. 2: Cartoon-block image generated by dssr_block.py for PDB entry 2n2d (an NMR ensemble).
In addition to the default settings, DSSR offers quite a few variations for the size and coloring of rectangular blocks, as demonstrated in Fig.3. The main settings are through the block_file option in PyMOL (note the underscore), corresponding to DSSR --block-file (or --block_file). The corresponding PyMOL commands are also listed for your reference. You can easily play around with the various styles interactively in PyMOL by toggling objects (dssr_block##) on or off. Enjoy!
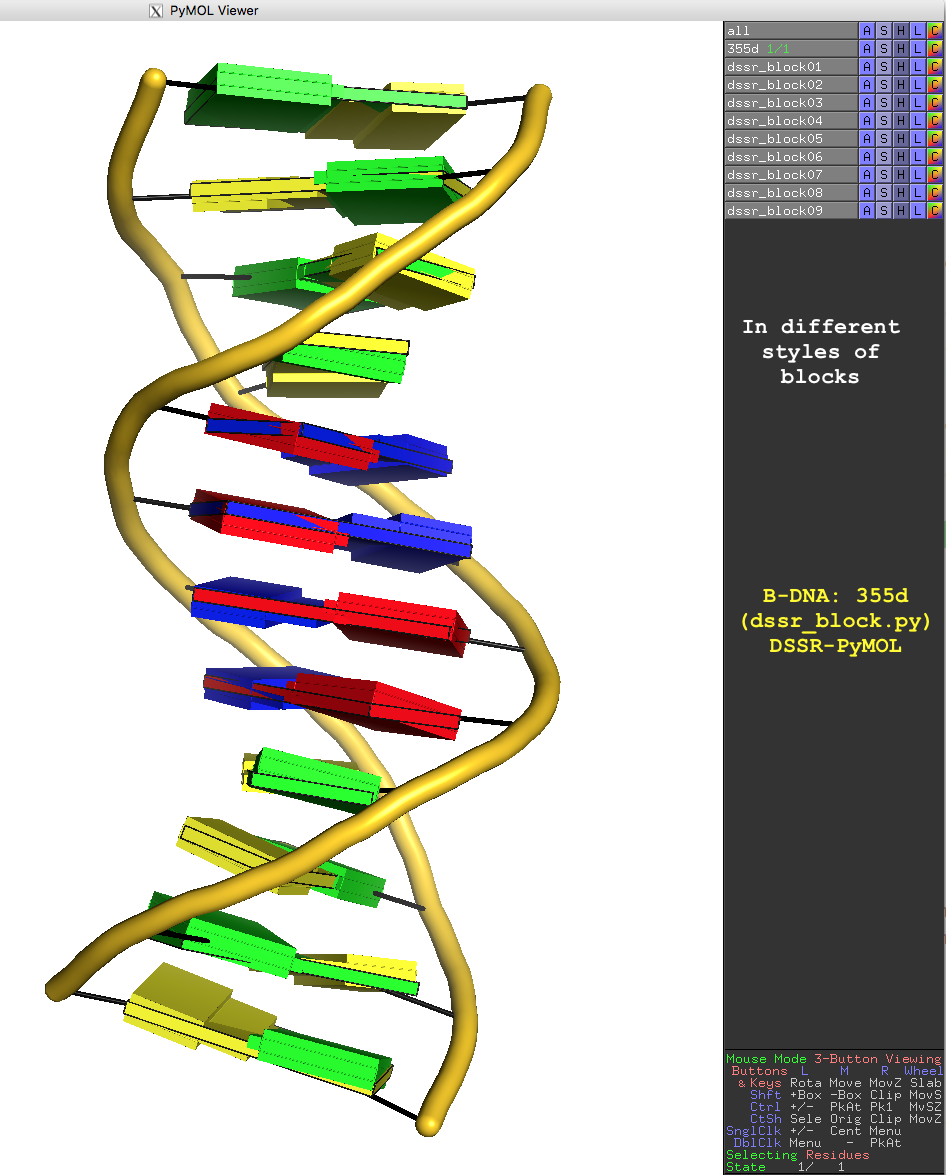
Fig. 3: Cartoon-block image generated by dssr_block.py for PDB entry 355d (the Dickerson B-DNA dodecamer).
Fig. 3 is created with the following PyMOL commands:
reinitialize fetch 355d, async=0 bg_color white as cartoon orient turn z, -90 turn y, 180 set cartoon_ladder_mode, 1 set cartoon_ladder_radius, 0.1 set cartoon_ladder_color, black set cartoon_tube_radius, 0.5 set cartoon_nucleic_acid_mode, 1 set cartoon_color, gold dssr_block 355d # default base blocks in solid color dssr_block block_file=edge # rectangular blocks in wireframe (black) dssr_block block_file=face+edge # solid color with outline dssr_block block_file=equal # bases blocks in equal size dssr_block block_file=minor # with minor-groove colord black dssr_block block_file=wc # Watson-Crick base pairs in long bp blocks dssr_block block_file=wc-minor # Watson-Crick pairs + minor-groove edge dssr_block block_file=gray # rectangular blocks all in gray dssr_block block_depth=1.8 # with increased thickness
Notes
- The
dssr_block.pyscript described here is the original version Thomas communicated to me. Current version of this script and related topics can be found in the Dssr block PyMOLWiki page. - For this script to work, DSSR needs to be installed and
x3dna-dssrin the PATH. - In PyMOL,
set cartoon_nucleic_acid_mode, 1employs C3′ instead of the default P (‘mode 0’) for the smooth backbone trace. Since 5′ terminal phosphate groups are normally not available from X-ray crystal structures (e.g., 355d), ‘mode 1’ is used to avoid orphan base blocks from the backbone trace.
