An abasic site is a location in DNA or RNA where a purine or pyrimidine base is missing. It is also termed an AP site (i.e., apurinic/apyrimidinic site) in biochemistry and molecular genetics. The abasic site can be formed either spontaneously (e.g., depurination) or due to DNA damage (occurring as intermediates in base excision repair). According to Wikipedia, “It has been estimated that under physiological conditions 10,000 apurinic sites and 500 apyrimidinic may be generated in a cell daily.”
In DSSR and 3DNA v2.x, nucleotides are recognized using standard atom names and base planarity. Thus, abasic sites are not taken as nucleotides (by default), simply because they do not have base atoms. DSSR introduced the --abasic option to account for abasic sites, a feature useful for detecting loops with backbone connectivity.
For example, by default, DSSR identifies one internal loop (no. 1 in the list below) in PDB entry 1l2c. With the --abasic option, two internal loops (including the one with the abasic site C.HPD18, no. 2) are detected.
List of 2 internal loops
1 symmetric internal loop: nts=6; [1,1]; linked by [#-1,#1]
summary: [2] 1 1 [B.1 C.24 B.3 C.22] 1 4
nts=6 GTATAC B.DG1,B.DT2,B.DA3,C.DT22,C.DA23,C.DC24
nts=1 T B.DT2
nts=1 A C.DA23
2 symmetric internal loop: nts=6; [1,1]; linked by [#1,#2]
summary: [2] 1 1 [B.6 C.19 B.8 C.17] 4 5
nts=6 CTTA?G B.DC6,B.DT7,B.DT8,C.DA17,C.HPD18,C.DG19
nts=1 T B.DT7
nts=1 ? C.HPD18
Note that C.HPD18 in 1l2c is a non-standard residue, as shown in the HETATM records below. Since the identity of C.HPD18 cannot be deduced from the atomic records, its one-letter code is designated as ?.
HETATM 346 P HPD C 18 -14.637 52.299 29.949 1.00 49.12 P
HETATM 347 O5' HPD C 18 -14.658 52.173 28.359 1.00 48.28 O
HETATM 348 O1P HPD C 18 -15.167 51.040 30.537 1.00 49.35 O
HETATM 349 O2P HPD C 18 -13.303 52.798 30.369 1.00 46.43 O
HETATM 350 C5' HPD C 18 -15.703 51.469 27.687 1.00 45.70 C
HETATM 351 O4' HPD C 18 -16.364 50.501 25.561 1.00 44.15 O
HETATM 352 O3' HPD C 18 -13.990 51.738 24.335 1.00 45.75 O
HETATM 353 C1' HPD C 18 -16.105 54.187 25.684 1.00 52.47 C
HETATM 354 O1' HPD C 18 -17.309 54.085 26.496 1.00 56.16 O
HETATM 355 C3' HPD C 18 -14.756 52.250 25.426 1.00 46.23 C
HETATM 356 C4' HPD C 18 -15.263 51.093 26.291 1.00 45.72 C
HETATM 357 C2' HPD C 18 -16.030 52.889 24.898 1.00 49.05 C
In contrast, the R.U-8 in PDB entry 4ifd is a standard U, and is properly labeled by DSSR.
ATOM 26418 P U R -8 139.362 21.962 129.430 1.00208.29 P
ATOM 26419 OP1 U R -8 140.062 20.821 130.074 1.00207.30 O
ATOM 26420 OP2 U R -8 140.113 23.208 129.129 1.00208.44 O1+
ATOM 26421 O5' U R -8 138.712 21.439 128.071 1.00157.60 O
ATOM 26422 C5' U R -8 139.507 20.790 127.087 1.00155.47 C
ATOM 26423 C4' U R -8 138.843 20.804 125.731 1.00152.27 C
ATOM 26424 O4' U R -8 138.538 22.172 125.352 1.00149.29 O
ATOM 26425 C3' U R -8 139.677 20.275 124.572 1.00152.70 C
ATOM 26426 O3' U R -8 139.670 18.859 124.478 1.00155.04 O
ATOM 26427 C2' U R -8 139.053 20.969 123.369 1.00150.26 C
ATOM 26428 O2' U R -8 137.849 20.322 122.984 1.00146.83 O
ATOM 26429 C1' U R -8 138.700 22.334 123.958 1.00147.35 C
This is yet another little detail that DSSR takes care of. It is the close consideration to many such subtle points that makes DSSR different. Overall, DSSR represents my view of what a scientific software program could be (or should be).

Recently, while analyzing a representative set of RNA structures from the PDB, I came across three weird entries. They are documented below, primarily for my own record.
- 5els — “Structure of the KH domain of T-STAR in complex with AAAUAA RNA”. There are two alternative conformations for the six-nt
AAAUAA RNA component, labeled A and B, respectively. Normally, the A/B alternative coordinates for each atom are put directly next to each other, and assigned the same chain id, as in 1msy for the phosphate group of G2669 on chain A. In 5els, however, the two alternative conformations (A/B) are separated into two chains: chain H for A, and chain I for B.
- 1vql — “The structure of the transition state analogue ‘DCSN’ bound to the large ribosomal subunit of Haloarcula marismortui”. The three-nt fragment DA179—C180—C181 on chain 4 is in the 3’—>5’ direction.
- 4r3i — “The crystal structure of m(6)A RNA with the YTHDC1 YTH domain”. The mmCIF file has a model number of 0, instead of 1 (as in other cases I am aware of).

3DNA contains 55 fiber models compiled from literature, plus a derived RNA model (as of v2.1). To the best of my knowledge, this is the most comprehensive collection of regular DNA/RNA models. Please see Table 4 of the 2003 3DNA NAR paper for detailed structural features of these models and references.
The 55 models are based on the following works:
- Chandrasekaran & Arnott (from #1 to #43) — the most well-known set of fiber models
- Alexeev et al. (#44-#45)
- van Dam & Levitt (#46-#47)
- Premilat & Albiser (#48-#55)
The utility program fiber makes the generation of all these fiber models in a simple, consistent interface, and produces coordinate files in either PDB or PDBML format. Of those models, some can be built with an arbitrary sequence of A, C, G and T (e.g., A-/B-/C-DNA from calf thymus), while others are of fixed sequences (e.g., Z-DNA with GC repeats). The sequence can be specified either from command-line or a plain text file, in either lower, UPPER, or MixED cases.
Once 3DNA in properly installed, the command-line interface is the most versatile and convenient way to generate, e.g., a regular double-stranded DNA (mostly, B-DNA) of arbitrary sequence. The command-help message (generated with fiber -h) is as below:
NAME
fiber - generate 55 fiber models based on Arnott and other's work
SYNOPSIS
fiber [OPTION] PDBFILE
DESCRIPTION
generate 55 fiber models based on the repeating unit from Arnott's
work, including the canonical A-, B-, C- and Z-DNA, triplex, etc
-xml output structure coordinates in PDBML format
-num a structure identification number in the range (1-55)
-m, -l brief description of the 55 fiber structures
-a, -1 A-DNA model (calf thymus)
-b, -4 B-DNA (calf thymus, default)
-c, -47 C-DNA (BII-type nucleotides)
-d, -48 D(A)-DNA ploy d(AT) : ploy d(AT) (right-handed)
-z, -15 Z-DNA poly d(GC) : poly d(GC)
-rna for RNA with arbitrary base sequence
-seq=string specifying an arbitrary base sequence
-single output a single-stranded structure
-h this help message (any non-recognized options will do)
INPUT
An structural identification number (symbol)
EXAMPLES
fiber fiber-BDNA.pdb
# fiber -4 fiber-BDNA.pdb
# fiber -b fiber-BDNA.pdb
fiber -a fiber-ADNA.pdb
fiber -seq=AAAGGUUU -rna fiber-RNA.pdb
fiber -seq=AAAGGUUU -rna -single fiber-ssRNA.pdb
OUTPUT
PDB file
SEE ALSO
analyze, anyhelix, find_pair
AUTHOR
3DNA v2.3-2016sept06, created and maintained by Xiang-Jun Lu (PhD)
Please post questions/comments on the 3DNA Forum: http://forum.x3dna.org/
Moreover, the w3DNA, 3D-DART web-interfaces, and the PyMOL wrapper make it easy to generate a regular DNA (or RNA) model, especially for occasional users or for educational purposes.
In principle, nothing is worth showing off with regard to 3DNA’s fiber model generation functionality. Nevertheless, this handy tool serves as a clear example of the differences between a “proof of concept” and a pragmatic software application. I initially decided to work on this tool simply for my own convenience. At that time, I had access to A-DNA and B-DNA fiber model generators, each as a separate program. Moreover, the constructed models did not comply to the PDB format in atom naming, among other subtitles.
I started with the Chandrasekaran & Arnott fiber models which I had a copy of data files. However, there were many details to work out, typos to correct, etc. to put them in a consistent framework. For other models, I had to read each original publication, and to type raw atomic cylindrical coordinates into computer. Again, quite a few inconsistencies popped up between the different publications with a time span over decades.
Overall, it was a quite tedious undertaking, requiring great attention to details. I am glad that I did that: I learned so much from the process, and more importantly, others can benefit from my effort. As I put in the 3DNA Nature Protocol paper (BOX 6 | FIBER-DIFFRACTION MODELS),
In preparing this set of fiber models, we have taken great care to ensure the accuracy and consistency of the models. For completeness and user verification, 3DNA includes, in addition to 3DNA-processed files, the original coordinates collected from the literature.
For those who want to understand what’s going on under the hood, there is no better way than to try to reproduce the process using, e.g., fiber B-DNA as an example.
From the very beginning, I had expected the 3DNA fiber functionality to serve as a handy tool for building a regular DNA duplex of chosen sequence. Over the years, the fiber program has gradually attracted attention from the community. The recent PyMOL wrapper by Thomas Holder is a clear sign of its increased popularity, and has prompted me to write this post, adapted largely from the one titled Fiber models in 3DNA make it easy to build regular DNA helices (dated Friday, October 9, 2009).
See also PyMOL wrapper to 3DNA fiber models

Given below is the content of the README file for fiber models in 3DNA:
1. The repeating units of each fiber structure are mostly based on the
work of Chandrasekaran & Arnott (from #1 to #43). More recent fiber
models are based on Alexeev et al. (#44-#45), van Dam & Levitt (#46
-#47) and Premilat & Albiser (#48-#55).
2. Clean up of each residue
a. currently ignore hydrogen atoms [can be easily added]
b. change ME/C7 group of thymine to C5M
c. re-assign O3' atom to be attached with C3'
d. change distance unit from nm to A [most of the entries]
e. re-ordering atoms according to the NDB convention
3. Fix up of problem structures.
a. str#8 has no N9 atom for guanine
b. str#10 is not available from the disk, manually input
c. str#14 C5M atom was named C5 for Thymine, resulting two C5 atoms
d. str#17 has wrong assignment of O3' atom on Guanine
e. str#33 has wrong C6 position in U3
f. str#37 to #str41 were typed in manually following Arnott's
new list as given in "Oxford Handbook of Nucleic Acid Structure"
edited by S. Neidle (Oxford Press, 1999)
g. str#38 coordinates for N6(A) and N3(T) are WRONG as given in the
original literature
h. str#39 and #40 have the same O3' coordinates for the 2nd strand
4. str#44 & 45 have fixed strand II residues (T)
5. str#46 & 47 have +z-axis upwards (based on BI.pdb & BII.pdb)
6. str#48 to 55 have +z-axis upwards
List of 55 fiber structures
id# Twist Rise Structure description
(dgrees) (A)
-------------------------------------------------------------------------------
1 32.7 2.548 A-DNA (calf thymus; generic sequence: A, C, G and T)
2 65.5 5.095 A-DNA poly d(ABr5U) : poly d(ABr5U)
3 0.0 28.030 A-DNA (calf thymus) poly d(A1T2C3G4G5A6A7T8G9G10T11) :
poly d(A1C2C3A4T5T6C7C8G9A10T11)
4 36.0 3.375 B-DNA (calf thymus; generic sequence: A, C, G and T)
5 72.0 6.720 B-DNA poly d(CG) : poly d(CG)
6 180.0 16.864 B-DNA (calf thymus) poly d(C1C2C3C4C5) : poly d(G6G7G8G9G10)
7 38.6 3.310 C-DNA (calf thymus; generic sequence: A, C, G and T)
8 40.0 3.312 C-DNA poly d(GGT) : poly d(ACC)
9 120.0 9.937 C-DNA poly d(G1G2T3) : poly d(A4C5C6)
10 80.0 6.467 C-DNA poly d(AG) : poly d(CT)
11 80.0 6.467 C-DNA poly d(A1G2) : poly d(C3T4)
12 45.0 3.013 D-DNA poly d(AAT) : poly d(ATT)
13 90.0 6.125 D-DNA poly d(CI) : poly d(CI)
14 -90.0 18.500 D-DNA poly d(A1T2A3T4A5T6) : poly d(A1T2A3T4A5T6)
15 -60.0 7.250 Z-DNA poly d(GC) : poly d(GC)
16 -51.4 7.571 Z-DNA poly d(As4T) : poly d(As4T)
17 0.0 10.200 L-DNA (calf thymus) poly d(GC) : poly d(GC)
18 36.0 3.230 B'-DNA alpha poly d(A) : poly d(T) (H-DNA)
19 36.0 3.233 B'-DNA beta2 poly d(A) : poly d(T) (H-DNA beta)
20 32.7 2.812 A-RNA poly (A) : poly (U)
21 30.0 3.000 A'-RNA poly (I) : poly (C)
22 32.7 2.560 Hybrid poly (A) : poly d(T)
23 32.0 2.780 Hybrid poly d(G) : poly (C)
24 36.0 3.130 Hybrid poly d(I) : poly (C)
25 32.7 3.060 Hybrid poly d(A) : poly (U)
26 36.0 3.010 10-fold poly (X) : poly (X)
27 32.7 2.518 11-fold poly (X) : poly (X)
28 32.7 2.596 Poly (s2U) : poly (s2U) (symmetric base-pair)
29 32.7 2.596 Poly (s2U) : poly (s2U) (asymmetric base-pair)
30 32.7 3.160 Poly d(C) : poly d(I) : poly d(C)
31 30.0 3.260 Poly d(T) : poly d(A) : poly d(T)
32 32.7 3.040 Poly (U) : poly (A) : poly(U) (11-fold)
33 30.0 3.040 Poly (U) : poly (A) : poly(U) (12-fold)
34 30.0 3.290 Poly (I) : poly (A) : poly(I)
35 31.3 3.410 Poly (I) : poly (I) : poly(I) : poly(I)
36 60.0 3.155 Poly (C) or poly (mC) or poly (eC)
37 36.0 3.200 B'-DNA beta2 Poly d(A) : poly d(U)
38 36.0 3.240 B'-DNA beta1 Poly d(A) : poly d(T)
39 72.0 6.480 B'-DNA beta2 Poly d(AI) : poly d(CT)
40 72.0 6.460 B'-DNA beta1 Poly d(AI) : poly d(CT)
41 144.0 13.540 B'-DNA Poly d(AATT) : poly d(AATT)
42 32.7 3.040 Poly(U) : poly d(A) : poly(U) [cf. #32]
43 36.0 3.200 Beta Poly d(A) : Poly d(U) [cf. #37]
44 36.0 3.233 Poly d(A) : poly d(T) (Ca salt)
45 36.0 3.233 Poly d(A) : poly d(T) (Na salt)
46 36.0 3.38 B-DNA (BI-type nucleotides; generic sequence: A, C, G and T)
47 40.0 3.32 C-DNA (BII-type nucleotides; generic sequence: A, C, G and T)
48 87.8 6.02 D(A)-DNA ploy d(AT) : ploy d(AT) (right-handed)
49 60.0 7.20 S-DNA ploy d(CG) : poly d(CG) (C_BG_A, right-handed)
50 60.0 7.20 S-DNA ploy d(GC) : poly d(GC) (C_AG_B, right-handed)
51 31.6 3.22 B*-DNA poly d(A) : poly d(T)
52 90.0 6.06 D(B)-DNA poly d(AT) : poly d(AT) [cf. #48]
53 -38.7 3.29 C-DNA (generic sequence: A, C, G and T) (depreciated)
54 32.73 2.56 A-DNA (generic sequence: A, C, G and T) [cf. #1]
55 36.0 3.39 B-DNA (generic sequence: A, C, G and T) [cf. #4]
-------------------------------------------------------------------------------
List 1-41 based on Struther Arnott: ``Polynucleotide secondary structures:
an historical perspective'', pp. 1-38 in ``Oxford Handbook of Nucleic
Acid Structure'' edited by Stephen Neidle (Oxford Press, 1999).
#42 and #43 are from Chandrasekaran & Arnott: "The Structures of DNA
and RNA Helices in Oriented Fibers", pp 31-170 in "Landolt-Bornstein
Numerical Data and Functional Relationships in Science and Technology"
edited by W. Saenger (Springer-Verlag, 1990).
#44-#45 based on Alexeev et al., ``The structure of poly(dA) . poly(dT)
as revealed by an X-ray fiber diffraction''. J. Biomol. Str. Dyn, 4,
pp. 989-1011, 1987.
#46-#47 based on van Dam & Levitt, ``BII nucleotides in the B and C forms
of natural-sequence polymeric DNA: a new model for the C form of DNA''.
J. Mol. Biol., 304, pp. 541-561, 2000.
#48-#55 based on Premilat & Albiser, ``A new D-DNA form of poly(dA-dT) .
poly(dA-dT): an A-DNA type structure with reversed Hoogsteen Pairing''.
Eur. Biophys. J., 30, pp. 404-410, 2001 (and several other publications).

Recently, I heard from Thomas Holder, the PyMOL Principal Developer (Schrödinger, Inc.), that he had written a wrapper to the 3DNA fiber command. This PyMOL wrapper is implemented as part of his versatile PSICO library (see the PyMOL Wiki page Psico for details), and exposes the 55 fiber models based on Arnott and other’s work to the wide PyMOL user community. Moreover, the wrapper can be accessed directly from PyMOL (without installing PSICO), as shown below with an example:
PyMOL> run https://raw.githubusercontent.com/speleo3/pymol-psico/master/psico/creating.py
PyMOL> fiber CTAGCG
The resulting fiber model is the default B-form DNA of calf thymus, with twist of 36.0° and rise of 3.375 Å (see figure below). Note that cases in base sequence do not matter, so fiber ctagcg or fiber CTAgcg will give the same result.
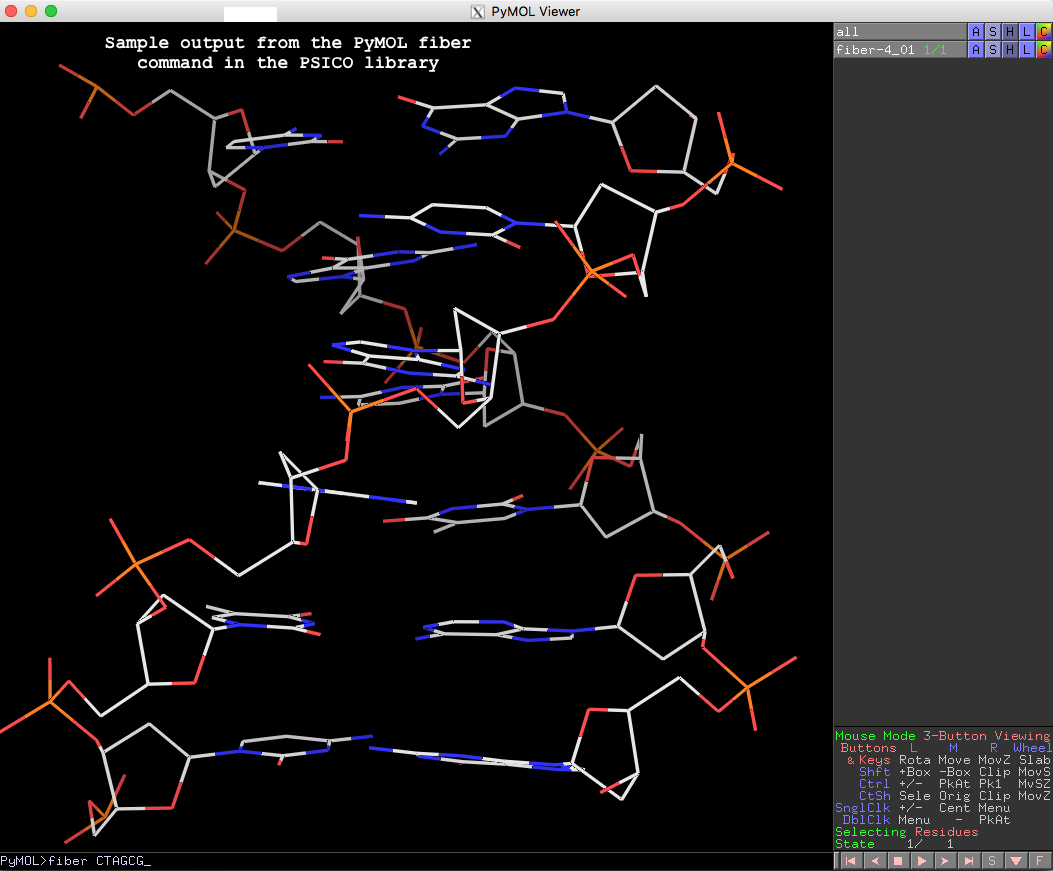
Running PyMOL>help fiber gives the following detailed usages info, which should be sufficient to get one started with this fiber tool in PyMOL.
PyMOL> help fiber
DESCRIPTION
Run X3DNA's "fiber" tool.
For the list of structure identification numbers, see for example:
http://xiang-jun.blogspot.com/2009/10/fiber-models-in-3dna.html
USAGE
fiber seq [, num [, name [, rna [, single ]]]]
ARGUMENTS
seq = str: single letter code sequence or number of repeats for
repeat models.
num = int: structure identification number {default: 4}
name = str: name of object to create {default: random unused name}
rna = 0/1: 0=DNA, 1=RNA {default: 0}
single = 0/1: 0=double stranded, 1=single stranded {default: 0}
EXAMPLES
# environment (this could go into ~/.pymolrc or ~/.bashrc)
os.environ["X3DNA"] = "/opt/x3dna-v2.3"
# B or A DNA from sequence
fiber CTAGCG
fiber CTAGCG, 1, ADNA
# double or single stranded RNA from sequence
fiber AAAGGU, name=dsRNA, rna=1
fiber AAAGGU, name=ssRNA, rna=1, single=1
# poly-GC Z-DNA repeat model with 10 repeats
fiber 10, 15
Thanks to Thomas, for making another connection between PyMOL and 3DNA/DSSR. The other one is the DSSR-plugin for PyMOL to create “block” shaped cartoons for nucleic acid bases and base pairs.
See also 3DNA fiber models

Recently I read the article titled Structural Insights into the Quadruplex−Duplex 3′ Interface Formed from a Telomeric Repeat: A Potential Molecular Target by Krauss et al.. I quickly ran DSSR on the corresponding PDB entry is 5dww. Not surprisingly, DSSR can automatically identify reported key structural features (see output file 5dww.out for details), including the TAT triplet at the quadruplex−duplex junction, and the three G-quartets. Note that the result is based on biological assembly 1 in PDB file 5dww.pdb1 since the asymmetric unit contains four such molecules.
List of 4 multiplets
1 nts=3 TAT 1:A.DT17,1:A.DA19,1:B.DT7
2 nts=4 GGGG 1:A.DG1,1:A.DG5,1:A.DG9,1:A.DG14
3 nts=4 GGGG 1:A.DG2,1:A.DG6,1:A.DG10,1:A.DG15
4 nts=4 GGGG 1:A.DG3,1:A.DG7,1:A.DG11,1:A.DG16
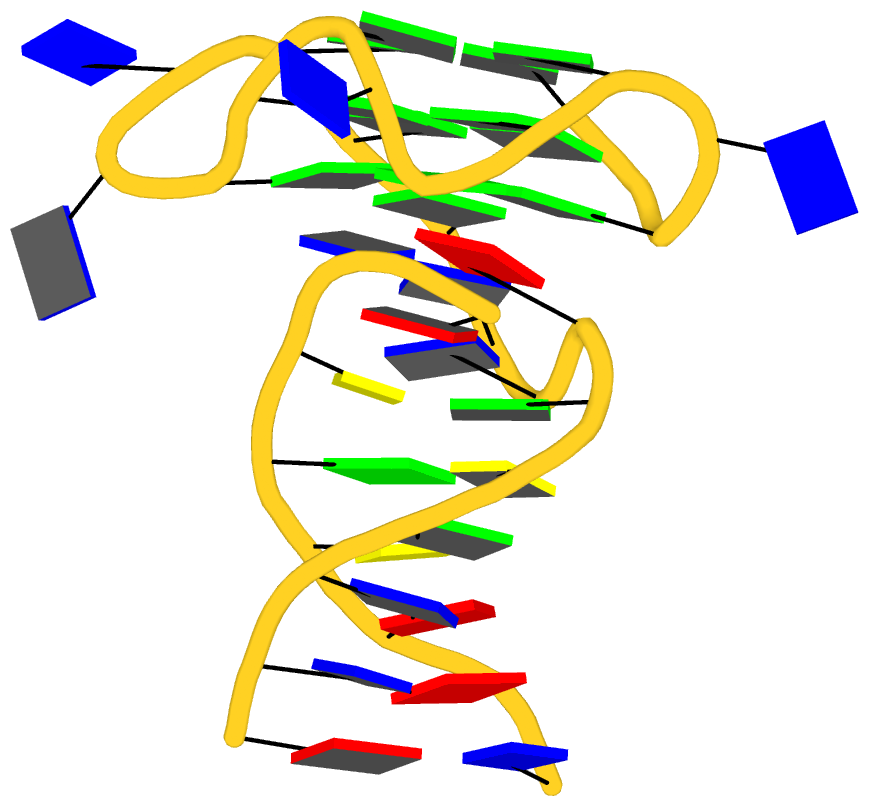
As its title suggests, however, this blog post is about the cartoon-block representations. Four styles of such schematics are shown below, which can all be easily generated using DSSR/PyMOL.
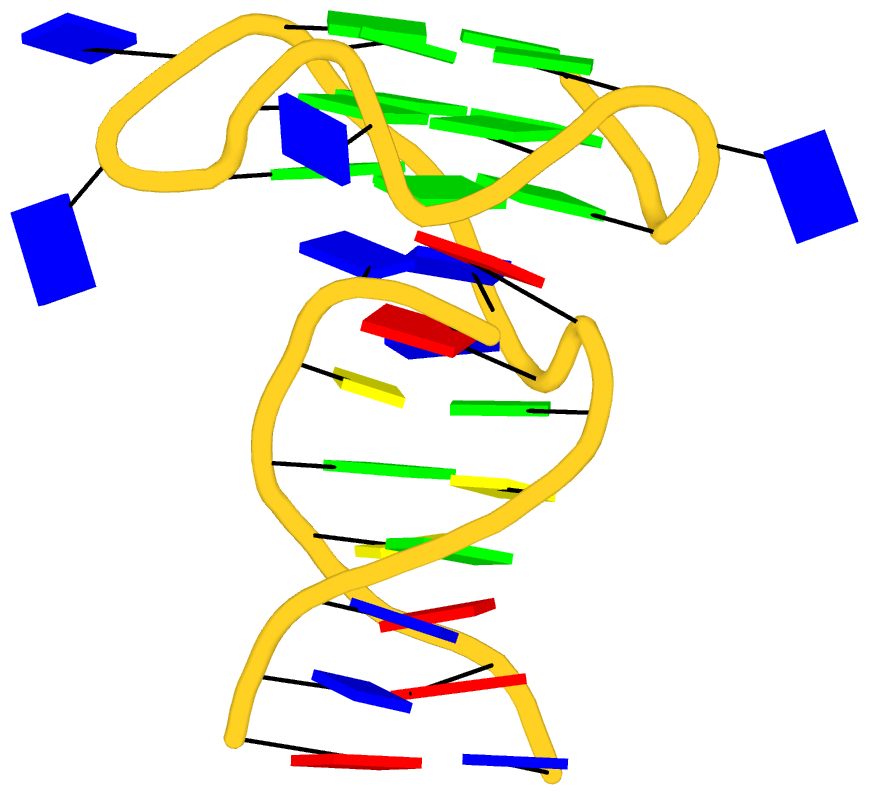 |
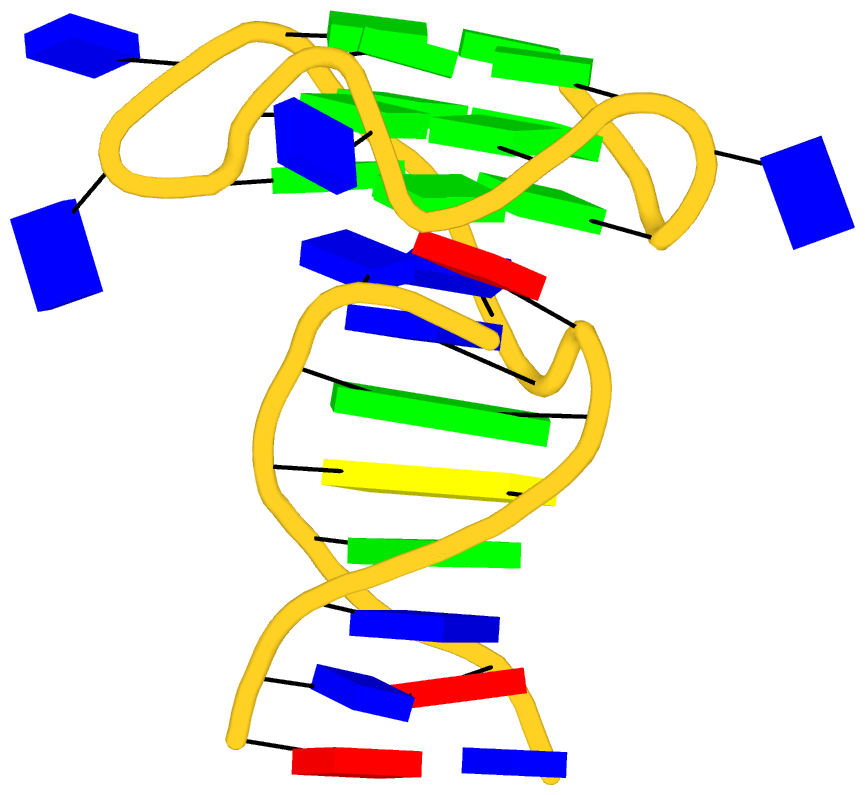 |
| in default style |
with base-pair blocks |
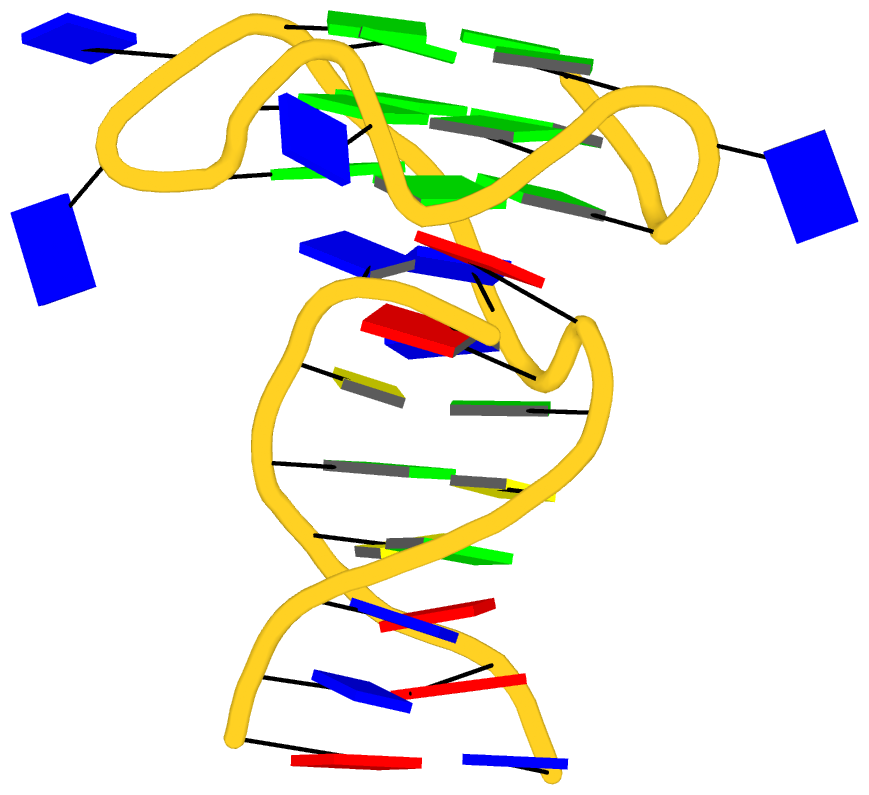 |
 |
| minor-groove highlighted |
top-face highlighted |
The cartoon-block representations possess unique features not seen elsewhere. With the help of the dssr_block in PyMOL, they are extremely easy to generate. Such schematics are likely to become popular in illustrations of nucleic acid structures.

As of today (2016-01-16), the number of registrations on the 3DNA Forum has reached 2,562. Moreover, all the members (as far as I can tell) are legitimate since the Forum has remained spam free. From the very beginning, ensuring a high information-to-noice ratio has been a top priority. The goal has been achieved by taking the following measures:
State the rules clearly in the “Registration Agreement”
This forum is dedicated to topics generally related to the 3DNA suite of software programs for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. To make the 3DNA forum a more pleasant virtual community for all of us to learn from and contribute to, please be considerate and practice good netiquette (http://www.albion.com/netiquette/).
I strive to make the forum spam free. Specifically, posts that are not 3DNA related in the broad sense are taken as spams, and are strictly forbidden. You are solely responsible for the content of your posts. We reserve the right to remove any post deemed as inappropriate, deactivate the account and ban the IP address of any abuser of the forum, WITHOUT NOTICE.
When posting on the Forum, please abide by the following rules: …
In a nutshell, you are welcome to participate and should not hesitate to ask questions, but remember to play nice and preferably share what you’ve learned! Please note that we do not tolerate spamming or off-topic trolling of any form.
Take advantage of anti-spam software
In additional to the verification of email address and check for black-listed IP addresses, the topic-specific questions have been very effective. Three examples of such questions are shown below:
What does the 'A' in 3DNA stand for? (hint: 4-char long)
How many standard bases does RNA have (hint: 1-digit number)
What is the value of the expression (3.1498 * 0 + 168)?
Overall, I do not like CAPTCHA — I’ve found the highly-distorted images in some websites especially troublesome. For the first few of years (to ~2014), the 3DNA Forum did not contain a captcha image in the registration page. Later on, however, I’ve noticed quite a few spam registrations/posts. In addition to quickly cleaning them up manually, I had refined the topic-specific questions, and turned on the visual verification image at level “Medium — Overlapping colored letters, with noise/lines”. Experience over the past couple of year has demonstrated the effectiveness of the combined strategy. As shown in the screen capture below, as of this writing, 177,562 spammers have been blocked by the anti-spam software!
Verify and approve ‘suspect’ accounts quickly
The above mentioned anti-spaming measures have blocked virtually all the “bad guys” so I do not need to waste time fighting them. I receive an email notification for each successful registration. The vast majority of registrants can then immediately access the member-only download section or post questions on the 3DNA Forum after registration. A significant portion (~1 out of 6) of the registrations, however, would be masked as suspicious and need my action. The email message for such cases reads like this:
‘xxxx’ has just signed up as a new member of your forum. Click the link below to view their profile. …
Before this member can begin posting they must first have their account approved. Click the link below to go to the approval screen. …
Wherever I have access to the Internet (including after hours with an iPad Air 2), I’ve always been quick in verifying and (mostly) approving these registrations.
Overall, since http://forum.x3dna.org was created in December 2011, the Forum has received significant attention in the field of DNA/RNA structural bioinformatics. As the community begins to appreciate and fully take advantage of what DSSR and SNAP have to offer, I have no doubt the Forum will gain even wider-spread recognition.

In recent years, reproducibility of ‘scientific’ publications has become quite a topic. See a recent essay Five selfish reasons to work reproducibly by Markowetz in Genome Biology (2015, 16:274). There are numerous reasons why reproducibility could become an issue at all in science. What I have continuously strived for in my scientific career, however, is to ensure that my published results are reproducible. As a concrete example, I created a dedicated section titled DSSR-NAR paper on the 3DNA Forum that provides full details (scripts and data files) so that any interested parties can rigorously reproduce the results reported in the DSSR Nucleic Acids Research (NAR) paper.
In my support of 3DNA for over a decade, the #1 issue I experienced is undoubtedly vague (non-reproducible) questions. For example, I have recently been asked via email why the 3DNA find_pair/analyze programs miss “some basepair … even though it is in the pdb file”. Without access to the PDB file to reproduce the problem, however, I cannot provide a concrete answer. In an effect to prevent ambiguous questions, I made the following explicit point in the “Registration Agreement” of the 3DNA Forum (no. 2 on the list):
Be specific with your questions; provide a minimal, reproducible example if possible; use attachments where appropriate.
The #2 issue is receiving 3DNA-related questions privately instead of on the intended public 3DNA Forum. I turned off “personal messaging” to receive private messages on the Forum long time ago, yet I have kept receiving questions via emails. In several locations on the 3DNA Forum, I have made this ‘public-question’ policy crystal clear:
Ask your questions in the public 3DNA forum instead of sending xiangjun emails or personal messages. (no. 1 on the ‘Registration Agreement’)
Please be aware that for the benefit of the 3DNA-user community at large, I do not provide private email/personal message support; the forum has been created specifically for open discussions of all 3DNA-related issues. In other words, any 3DNA-associated questions are welcome and should be directed here. Presumably I’ve made the message simple and clear enough to get across without further explanation. (in ‘Site announcements » Download instructions’ and ‘Downloads » 3DNA download’)
In response to the many 3DNA-related questions that still keep coming via email, I created the following entry of Canned Responses in gmail:
Thanks for your interest in using 3DNA. Please be aware that for the benefit of the 3DNA-user community at large, I do not provide private email support; the 3DNA Forum (http://forum.x3dna.org/) has been created specifically for open discussions of all 3DNA-related issues. In other words, any 3DNA-associated questions are welcome and should be directed there. I monitor the forum regularly and respond to posts promptly.
I look forward to seeing you on the 3DNA Forum (http://forum.x3dna.org/).
Overall, I’ve learned from experience that addressing reproducible questions publicly does the best for the 3DNA community. Users can register with personal (free) email address, and post simulated data to illustrate the problem at hand. Moreover, questions on the Forum have always received quick responses. Over time, the Forum has served as an archive that everyone can benefit from.

As of v2.3-2016jan01, the 3DNA analyze program outputs a list of new ‘simple’ base-pair and step parameters, by default. Shown below is a sample output for PDB entry 1xvk. This echinomycin-(GCGTACGC)2 complex has a single DNA strand as the asymmetric unit. 3DNA needs the the biological unit (1xvk.pdb1) to analyze the duplex (with the -symm option). This structure contains two Hoogsteen base pairs, and has popped up on the 3DNA Forum for the zero or negative Rise values. Note that the ‘simple’ Rise values are all positive; for the middle (#4) TA/TA step, it is now 3.09 Å instead of 0.
# find_pair -symm 1xvk.pdb1 1xvk.bps
# analyze -symm 1xvk.bps
# OR by combing the above two commands:
# find_pair -symm 1xvk.pdb1 | analyze -symm
# The output is in file '1xvk.out'
This structure contains 4 non-Watson-Crick (with leading *) base pair(s)
----------------------------------------------------------------------------
Simple base-pair parameters based on RC8--YC6 vectors
bp Shear Stretch Stagger Buckle Propeller Opening
* 1 G+C -3.07 1.55 -0.35 -6.98 0.29 67.33
2 C-G 0.27 -0.17 0.35 -22.34 3.33 -2.80
3 G-C -0.39 -0.17 0.41 22.91 1.81 -2.73
* 4 T+A -3.29 1.56 0.31 -8.03 1.59 -70.46
* 5 A+T -3.29 1.56 -0.31 -8.03 1.59 70.46
6 C-G 0.39 -0.17 0.41 -22.91 1.81 -2.72
7 G-C -0.27 -0.17 0.35 22.34 3.32 -2.80
* 8 C+G -3.07 1.55 0.35 -6.98 0.30 -67.33
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
ave. -1.59 0.69 0.19 -3.75 1.75 -1.38
s.d. 1.72 0.92 0.32 17.57 1.15 52.11
----------------------------------------------------------------------------
Simple base-pair step parameters based on consecutive C1'-C1' vectors
step Shift Slide Rise Tilt Roll Twist
* 1 GC/GC -0.55 0.39 7.41 6.40 -4.22 23.36
2 CG/CG -0.05 0.87 2.44 -0.55 3.94 -0.81
* 3 GT/AC 0.38 0.47 7.23 -8.62 3.75 25.70
* 4 TA/TA -0.00 4.73 3.09 -0.00 7.49 25.67
* 5 AC/GT -0.38 0.47 7.23 8.62 3.75 25.70
6 CG/CG 0.05 0.87 2.44 0.55 3.94 -0.82
* 7 GC/GC 0.55 0.39 7.41 -6.40 -4.22 23.36
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
ave. -0.00 1.17 5.32 -0.00 2.06 17.45
s.d. 0.39 1.59 2.50 6.21 4.49 12.52
The simple parameters are ‘intuitive’ for non-Watson-Crick base pairs and associated base-pair steps, where the existing standard-reference-frame-based 3DNA parameters may look weird. Note that these simple parameters are for structural description only, not to be fed into the ‘rebuild’ program. Overall, they complement the rigorous characterization of base-pair geometry, as demonstrated by the original analyze/rebuild pair of programs in 3DNA.
In short, the ‘simple’ base-pair parameters employ the YC6—RC8 vector as the y-axis whereas the ‘simple’ step parameters use consecutive C1’—C1’ vectors. As before, the z-axis is the average of two base normals, taking consideration of the M–N vs M+N base-pair classification. In essence, the ‘simple’ parameters make geometrical sense by introducing an ad hoc base-pair reference frame in each case. More details will be provided in a series of blog posts shortly.
Overall, this new section of ‘simple’ parameters should be taken as experimental. The output can be turned off by specifying the analyze -simple=false command-line option explicitly. As always, I greatly appreciate your feedback.

Over the years, I have played quite a few computer programming languages. ANSI C has become my top choice for ‘serious’ software projects, due to its small size, efficiency, flexibility, and ubiquitous support. Moreover, C is a mature language, with a rich ecosystem. As it turns out, C has also been consistently rated as one of the most popular computer languages (#1 or #2) over the past thirty years.
Needless to say, ANSI C has its own quirks, and it takes a steep learning curve. However, once you get over the hurdles, the language serves you. I cannot remember when, but it has been a long while that coding in ANSI C is no longer an issue. It is the understanding of scientific questions that takes most of my time, and coding helps greatly in refining my thoughts.
Not surprisingly, ANSI C was chosen as the sole language for DSSR (and SNAP, or 3DNA in general). The ensure the overall quality of the DSSR codebase, I have taken the following steps:
- The whole project is under git.
- The ANSI C source code is compiled with strict GCC options for full compliance to the standard:
-ansi -pedantic -W -Wall -Wextra -Wunused -Wshadow -Werror -O3
- The executable is checked with valgrind for any memory leak:
valgrind --leak-check=full x3dna-dssr -i=1ehz.pdb -o=1ehz.out --quiet
==19624== Memcheck, a memory error detector
==19624== Copyright (C) 2002-2013, and GNU GPL'd, by Julian Seward et al.
==19624== Using Valgrind-3.10.1 and LibVEX; rerun with -h for copyright info
==19624== Command: x3dna-dssr -i=1ehz.pdb -o=1ehz.out --quiet
==19624==
==19624==
==19624== HEAP SUMMARY:
==19624== in use at exit: 0 bytes in 0 blocks
==19624== total heap usage: 52,829 allocs, 52,829 frees, 92,878,578 bytes allocated
==19624==
==19624== All heap blocks were freed -- no leaks are possible
==19624==
==19624== For counts of detected and suppressed errors, rerun with: -v
==19624== ERROR SUMMARY: 0 errors from 0 contexts (suppressed: 0 from 0)
- Extensive tests (with the simple
diff command) to ensure the program is working as expected.
The above four measures combined allow me to add new features, refactor the code, and fix bugs, without worrying about accidentally breaking existing functionality. Reading literature (including citations to 3DNA/DSSR) and responding to user feedback on the 3DNA Forum keep me continuously improve DSSR. Some of the recent refinements to DSSR came about this way.







