Recently, I came across the paper by Mitra et al. (2025) titled "RNAproDB: A Webserver and Interactive Database for Analyzing Protein-RNA Interactions." I am glad to notice that DSSR (Lu et al. 2015) has been cited extensively in this work, as follows:
As part of the processing pipeline, multiple software is run including DSSR^12^ (base-pairing geometries, protein–RNA hydrogen bonds, and RNA secondary structure), HBPLUS^17^ (hydrogen bonds involving water molecules), ... Leontis-Westhof^27^ base pair annotations (as computed by DSSR^12^) ... The structural elements (stems, loops, hairpins, junctions, etc.) are detected using DSSR^12^ and mapped to the partial projection layout (via averaging corresponding residue coordinates)... We explored the relative abundance of different standard nucleotides (A, C, G, and U) in helical vs. non-helical regions (as computed by DSSR^12^)...We quantified the propensity of base-pairing (as detected by DSSR^12^) between different RNA bases (Fig. 3D).
This is an impressive contribution on the characterization of protein-RNA interactions. Reading carefully through the paper and its supplemental PDF, I was intrigued by the following note on a water-mediated U-U base pair missed by DSSR.
Another important aspect to discuss is RNA–RNA water-mediated interactions^33,34^. ... One such example is the CUG repeat structure from PDB ID 7Y2B^35^ (Fig. S5A). The U/U mismatches in this structure are often unable to form direct hydrogen bonds (specifically, the central U/U mismatch forms no direct hydrogen bond). Therefore, DSSR^12^ does not classify it as a base pair. However, two water molecules form water-mediated hydrogen bonds between the two U bases. ...
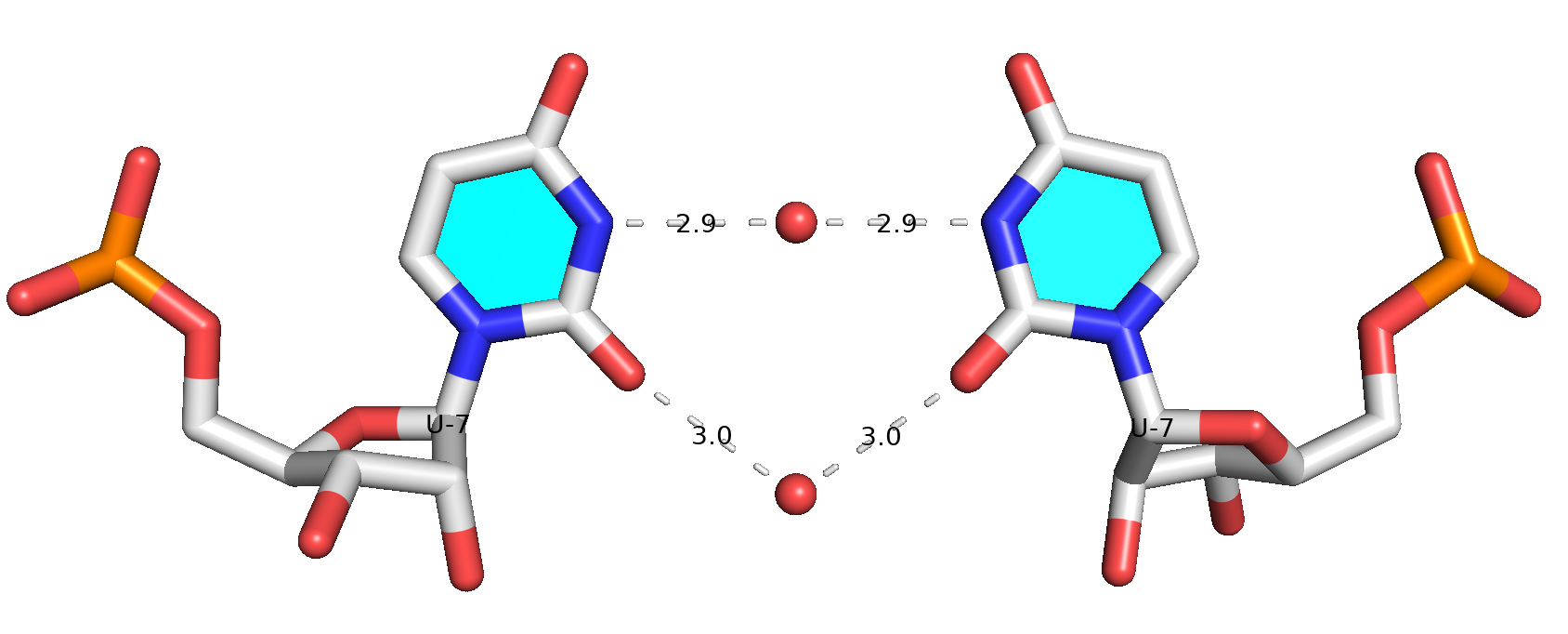
While DSSR internally already takes consideration of water-mediated H-bonds in the detection of base pairs, it still requires: (1) at least one direct H-bond between two base atoms or a base atom to backbone, and (2) a co-planar geometry between the two bases. The water-mediated U7-U7 pair in PDB entry 7Y2B does not fulfill condition (1): the minimal distance between the two U bases is 5 Å, which is far larger than a typical H-bonding distance. Therefore, DSSR did not classify it as a base pair.
Prompted by the observation of Mitra et al. (2025), I have added a new option (--pair-water) in the DSSR v2.5.1-2025mar19 release to allow for water-mediated base pairs to be detected. Using PDB entry 7Y2B as an example, the DSSR command and related base-pairs output are shown below.
# x3dna-dssr -i=7Y2B.pdb1 --symm --pair-water
List of 13 base pairs
nt1 nt2 bp name Saenger LW DSSR
1 1:S.U1 2:S.A13 U-A WC 20-XX cWW cW-W
2 1:S.U2 2:S.A12 U-A WC 20-XX cWW cW-W
3 1:S.C3 2:S.G11 C-G WC 19-XIX cWW cW-W
4 1:S.U4 2:S.U10 U-U -- -- cWW cW-W
5 1:S.G5 2:S.C9 G-C WC 19-XIX cWW cW-W
6 1:S.C6 2:S.G8 C-G WC 19-XIX cWW cW-W
7 1:S.U7 2:S.U7 U-U Water -- cWW cW-W
8 1:S.G8 2:S.C6 G-C WC 19-XIX cWW cW-W
9 1:S.C9 2:S.G5 C-G WC 19-XIX cWW cW-W
10 1:S.U10 2:S.U4 U-U -- -- cWW cW-W
11 1:S.G11 2:S.C3 G-C WC 19-XIX cWW cW-W
12 1:S.A12 2:S.U2 A-U WC 20-XX cWW cW-W
13 1:S.A13 2:S.U1 A-U WC 20-XX cWW cW-WBase pair #7 is water-mediated, as shown in the molecular image below. Note that .pdb1 means biological unit 1, and the option --symm reads the two symmetry-related structures in the MODEL/ENDMDL delineated ensemble as a single structure. See the DSSR User Manual for more details.
References
-
Lu, Xiang-Jun, Harmen J. Bussemaker, and Wilma K. Olson. 2015. “DSSR: An Integrated Software Tool for Dissecting the Spatial Structure of RNA.” Nucleic Acids Research, July, gkv716. https://doi.org/10.1093/nar/gkv716.
- Mitra, Raktim, Ari S. Cohen, Wei Yu Tang, Hirad Hosseini, Yongchan Hong, Helen M. Berman, and Remo Rohs. 2025. “RNAproDB: A Webserver and Interactive Database for Analyzing Protein-RNA Interactions.” Journal of Molecular Biology, February, 169012. https://doi.org/10.1016/j.jmb.2025.169012.
