Recently I read the article titled Structural Insights into the Quadruplex−Duplex 3′ Interface Formed from a Telomeric Repeat: A Potential Molecular Target by Krauss et al.. I quickly ran DSSR on the corresponding PDB entry is 5dww. Not surprisingly, DSSR can automatically identify reported key structural features (see output file 5dww.out for details), including the TAT triplet at the quadruplex−duplex junction, and the three G-quartets. Note that the result is based on biological assembly 1 in PDB file 5dww.pdb1 since the asymmetric unit contains four such molecules.
List of 4 multiplets 1 nts=3 TAT 1:A.DT17,1:A.DA19,1:B.DT7 2 nts=4 GGGG 1:A.DG1,1:A.DG5,1:A.DG9,1:A.DG14 3 nts=4 GGGG 1:A.DG2,1:A.DG6,1:A.DG10,1:A.DG15 4 nts=4 GGGG 1:A.DG3,1:A.DG7,1:A.DG11,1:A.DG16
As its title suggests, however, this blog post is about the cartoon-block representations. Four styles of such schematics are shown below, which can all be easily generated using DSSR/PyMOL.
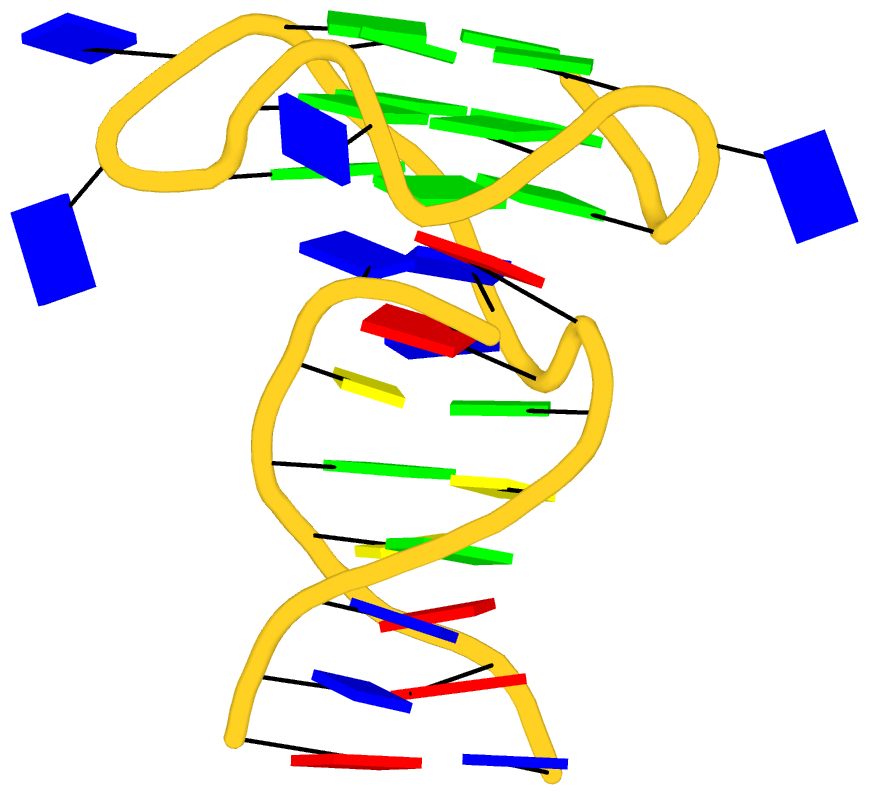 |
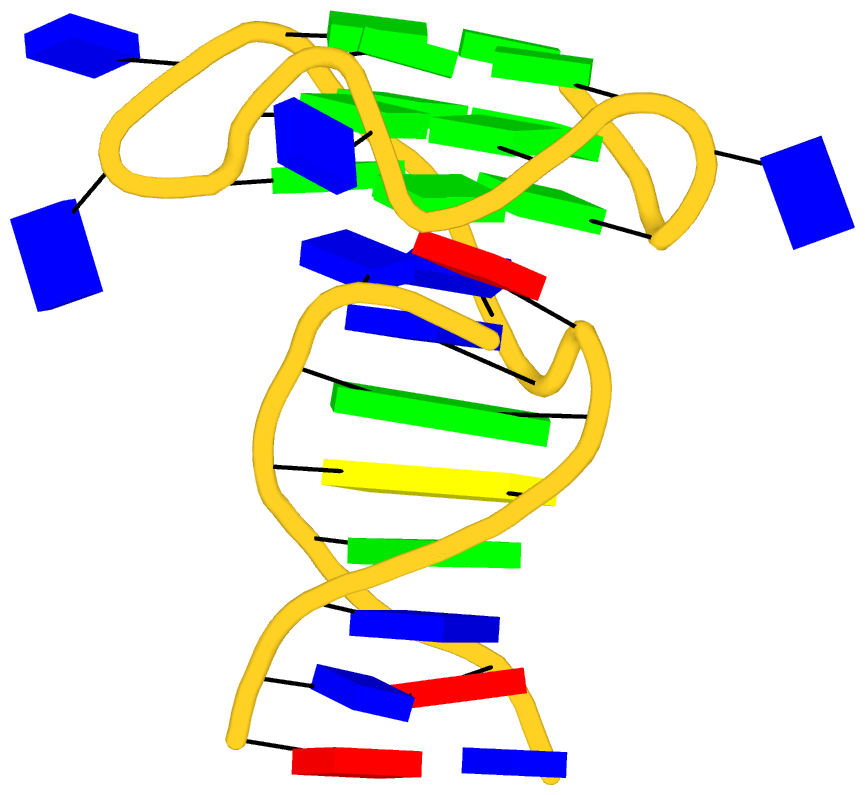 |
| in default style | with base-pair blocks |
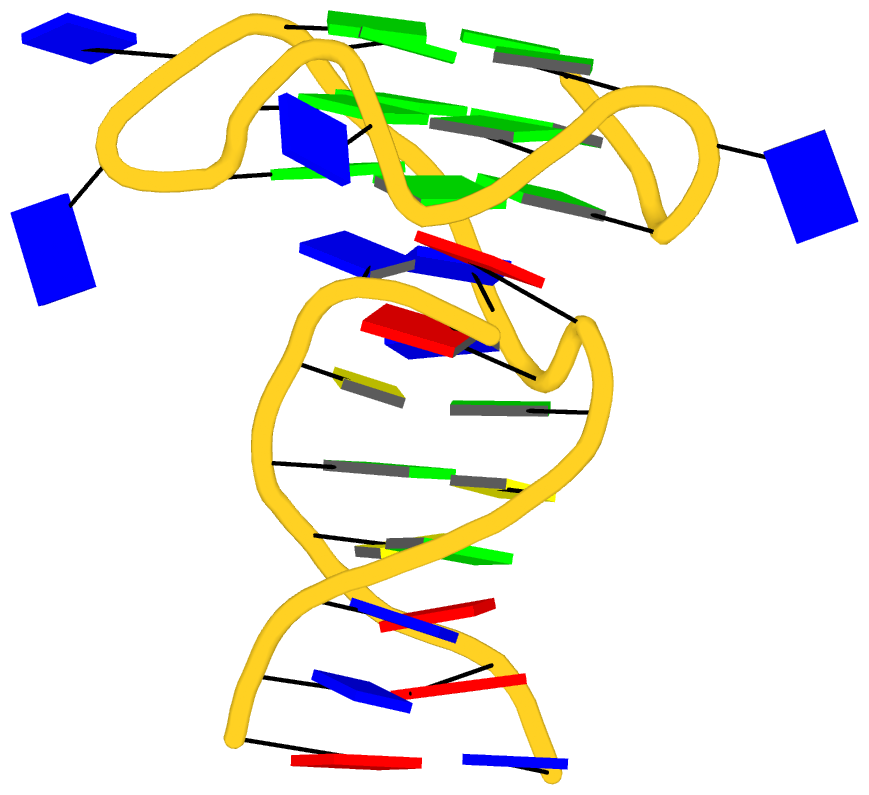 |
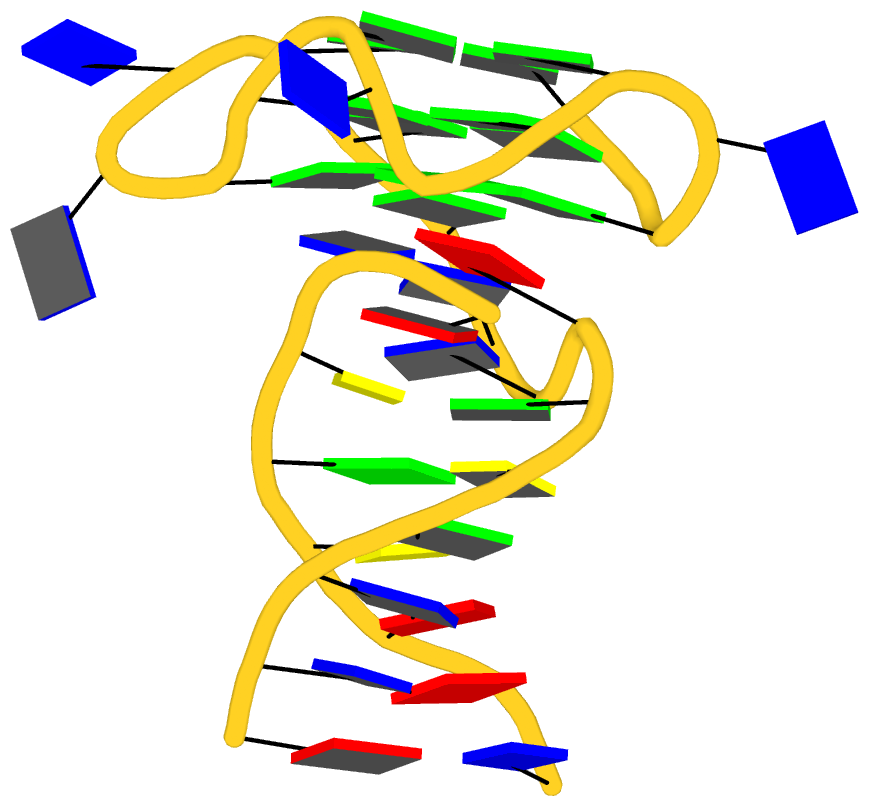 |
| minor-groove highlighted | top-face highlighted |
The cartoon-block representations possess unique features not seen elsewhere. With the help of the dssr_block in PyMOL, they are extremely easy to generate. Such schematics are likely to become popular in illustrations of nucleic acid structures.
