Recently, I heard from Thomas Holder, the PyMOL Principal Developer (Schrödinger, Inc.), that he had written a wrapper to the 3DNA fiber command. This PyMOL wrapper is implemented as part of his versatile PSICO library (see the PyMOL Wiki page Psico for details), and exposes the 55 fiber models based on Arnott and other’s work to the wide PyMOL user community. Moreover, the wrapper can be accessed directly from PyMOL (without installing PSICO), as shown below with an example:
PyMOL> run https://raw.githubusercontent.com/speleo3/pymol-psico/master/psico/creating.py PyMOL> fiber CTAGCG
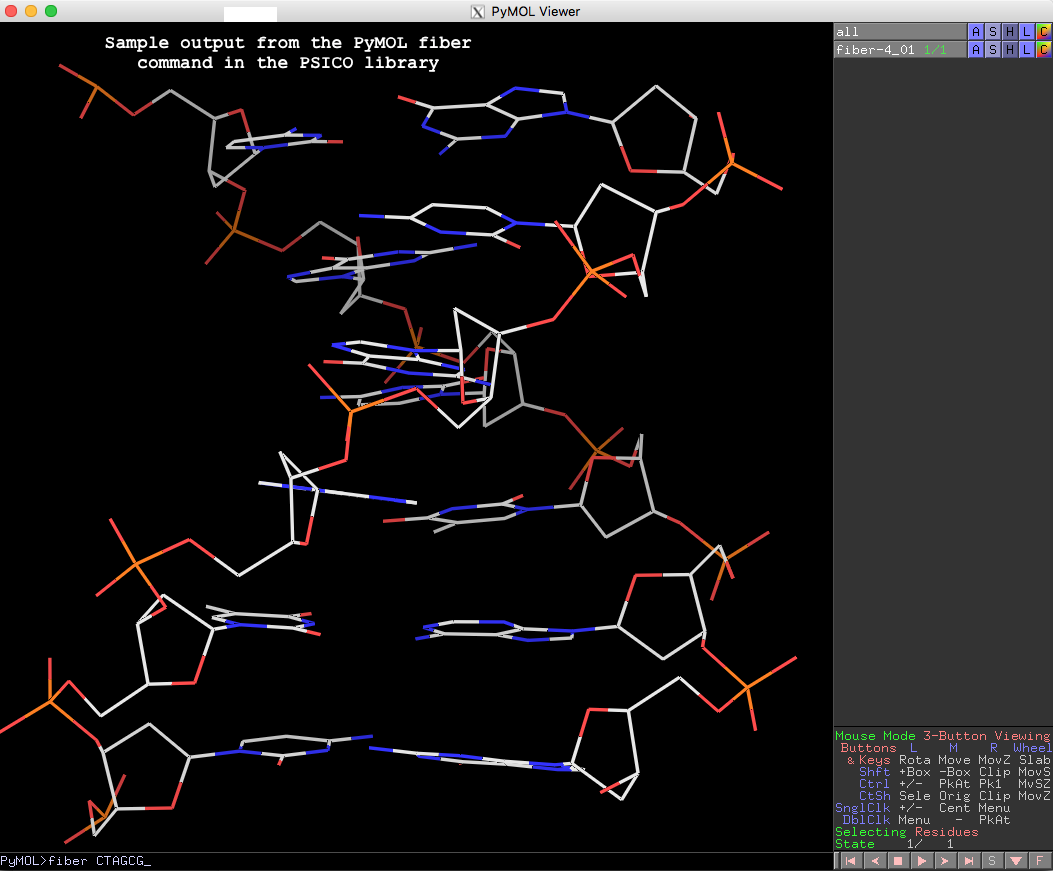
The resulting fiber model is the default B-form DNA of calf thymus, with twist of 36.0° and rise of 3.375 Å (see figure below). Note that cases in base sequence do not matter, so fiber ctagcg or fiber CTAgcg will give the same result.
Running PyMOL>help fiber gives the following detailed usages info, which should be sufficient to get one started with this fiber tool in PyMOL.
PyMOL> help fiber
DESCRIPTION
Run X3DNA's "fiber" tool.
For the list of structure identification numbers, see for example:
http://xiang-jun.blogspot.com/2009/10/fiber-models-in-3dna.html
USAGE
fiber seq [, num [, name [, rna [, single ]]]]
ARGUMENTS
seq = str: single letter code sequence or number of repeats for
repeat models.
num = int: structure identification number {default: 4}
name = str: name of object to create {default: random unused name}
rna = 0/1: 0=DNA, 1=RNA {default: 0}
single = 0/1: 0=double stranded, 1=single stranded {default: 0}
EXAMPLES
# environment (this could go into ~/.pymolrc or ~/.bashrc)
os.environ["X3DNA"] = "/opt/x3dna-v2.3"
# B or A DNA from sequence
fiber CTAGCG
fiber CTAGCG, 1, ADNA
# double or single stranded RNA from sequence
fiber AAAGGU, name=dsRNA, rna=1
fiber AAAGGU, name=ssRNA, rna=1, single=1
# poly-GC Z-DNA repeat model with 10 repeats
fiber 10, 15
Thanks to Thomas, for making another connection between PyMOL and 3DNA/DSSR. The other one is the DSSR-plugin for PyMOL to create “block” shaped cartoons for nucleic acid bases and base pairs.
See also 3DNA fiber models