Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. [Nucleic Acids Res 48: e74(https://doi.org/10.1093/nar/gkaa426)).
See the 2020 paper titled "DSSR-enabled innovative schematics of 3D nucleic acid structures with PyMOL" in Nucleic Acids Research and the corresponding Supplemental PDF for details. Many thanks to Drs. Wilma Olson and Cathy Lawson for their help in the preparation of the illustrations.
Details on how to reproduce the cover images are available on the 3DNA Forum.

Cryo-EM structure of the pre-B complex (PDB id: 8QP8; Zhang Z, Kumar V, Dybkov O, Will CL, Zhong J, Ludwig SE, Urlaub H, Kastner B, Stark H, Lührmann R. 2024. Structural insights into the cross-exon to cross-intron spliceosome switch. Nature 630: 1012–1019). The pre-B complex is thought to be critical in the regulation of splicing reactions. Its structure suggests how the cross-exon and cross-intron spliceosome assembly pathways converge. The U4, U5, and U6 snRNA backbones are depicted respectively by blue, green, and red ribbons, with bases and Watson-Crick base pairs shown as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; the proteins are represented by gold ribbons. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Structure of the Hendra henipavirus (HeV) nucleoprotein (N) protein-RNA double-ring assembly (PDB id: 8C4H; Passchier TC, White JB, Maskell DP, Byrne MJ, Ranson NA, Edwards TA, Barr JN. 2024. The cryoEM structure of the Hendra henipavirus nucleoprotein reveals insights into paramyxoviral nucleocapsid architectures. Sci Rep 14: 14099). The HeV N protein adopts a bi-lobed fold, where the N- and C-terminal globular domains are bisected by an RNA binding cleft. Neighboring N proteins assemble laterally and completely encapsidate the viral genomic and antigenomic RNAs. The two RNAs are depicted by green and red ribbons. The U bases of the poly(U) model are shown as cyan blocks. Proteins are represented as semitransparent gold ribbons. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Structure of the helicase and C-terminal domains of Dicer-related helicase-1 (DRH-1) bound to dsRNA (PDB id: 8T5S; Consalvo CD, Aderounmu AM, Donelick HM, Aruscavage PJ, Eckert DM, Shen PS, Bass BL. 2024. Caenorhabditis elegans Dicer acts with the RIG-I-like helicase DRH-1 and RDE-4 to cleave dsRNA. eLife 13: RP93979. Cryo-EM structures of Dicer-1 in complex with DRH-1, RNAi deficient-4 (RDE-4), and dsRNA provide mechanistic insights into how these three proteins cooperate in antiviral defense. The dsRNA backbone is depicted by green and red ribbons. The U-A pairs of the poly(A)·poly(U) model are shown as long rectangular cyan blocks, with minor-groove edges colored white. The ADP ligand is represented by a red block and the protein by a gold ribbon. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).
Moreover, the following 30 [12(2021) + 12(2022) + 6(2023)] cover images of the RNA Journal were generated by the NAKB (nakb.org).
Cover image provided by the Nucleic Acid Database (NDB)/Nucleic Acid Knowledgebase (NAKB; nakb.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).
It gives me great pleasure to announce that the 3DNA/DSSR project is now funded by the NIH R24GM153869 grant, titled "X3DNA-DSSR: a resource for structural bioinformatics of nucleic acids". I am deeply grateful for the opportunity to continue working on a project that has basically defined who I am. It was a tough time during the funding gap over the past few years. Nevertheless, I have experienced and learned a lot, and witnessed miracles enabled by enthusiastic users.
Since late 2020 when I lost my R01 grant, DSSR has been licensed by the Columbia Technology Ventures (CTV). I appreciate the numerous users (including big pharma) who purchased a DSSR Pro License or a DSSR Basic paid License. Thanks to the NIH R24GM153869 grant, we are pleased to provide DSSR Basic free of charge to the academic community. Academic Users may submit a license request for DSSR Basic or DSSR Pro by clicking "Express Licensing" on the CTV landing page. Commercial users may inquire about pricing and licensing terms by emailing techtransfer@columbia.edu, copying xiangjun@x3dna.org.
DSSR v2.4.5-2024sep24 was released to synchronize with the new R24 funding, which will bring the project to an entirely new level. All existing users are encouraged to upgrade their installation to this release which contains miscellaneous bug fixes (e.g., chain id with > 4 chars) and numerous minor improvements.
Lots of exciting things will happen for the project. The first thing is to make DSSR freely accessible to the academic community. In the past couple of weeks, CTV have already issued quite a few DSSR Basic Academic licenses to users from all over the world. So the demand is high, and it will become stronger as more academic users become aware of DSSR. I'm closely monitoring the 3DNA Forum, and is always ready to answer users questions.
I am committed to making DSSR a brand that stands for quality and value. By virtue of its unmatched functionality, usability, and support, DSSR saves users a substantial amount of time and effort when compared to other options. My track record throughout the years has unambiguously demonstrated my dedication to this solid software product.
DSSR Basic contains all features described in the three DSSR-related papers, and includes the originally separate SNAP program (still unpublished) for analyzing DNA/RNA-protein complexes. The Pro version integrates the classic 3DNA functionality, plus advanced modeling routines, with email/Zoom/phone support.
Recently, while analyzing a representative set of RNA structures from the PDB, I came across three weird entries. They are documented below, primarily for my own record.
- 5els — “Structure of the KH domain of T-STAR in complex with AAAUAA RNA”. There are two alternative conformations for the six-nt
AAAUAA RNA component, labeled A and B, respectively. Normally, the A/B alternative coordinates for each atom are put directly next to each other, and assigned the same chain id, as in 1msy for the phosphate group of G2669 on chain A. In 5els, however, the two alternative conformations (A/B) are separated into two chains: chain H for A, and chain I for B.
- 1vql — “The structure of the transition state analogue ‘DCSN’ bound to the large ribosomal subunit of Haloarcula marismortui”. The three-nt fragment DA179—C180—C181 on chain 4 is in the 3’—>5’ direction.
- 4r3i — “The crystal structure of m(6)A RNA with the YTHDC1 YTH domain”. The mmCIF file has a model number of 0, instead of 1 (as in other cases I am aware of).

Dear 3DNA Forum subscribers,
Here are some highlights of recent developments of 3DNA/DSSR:
Note: If you’ve difficulty in accessing the 3DNA homepage, possibly the case from mainland China (as I know it), please visit its duplicate at http://home.x3dna.org. This newsletter is written in Markdown, with a translated HTML version posted on the 3DNA homepage.
3DNA v2.3
The C source code is now available. Since the programs are written in strict ANSI C, 3DNA can be compiled (as is) on any computers/operating systems with a C (or C++) compiler. For user convenience, three binary distributions (with source code under the src/ subdirectory) are provided for Windows, Linux, and Mac OS X. The distributed Windows version works in native Windows (7 and up, via the cmd command-line interface, or ConEMU), MinGW/Msys (Msys2), and Cygwin, in either 32 or 64-bit.
A new set of ‘simple’ base-pair and step parameters was introduced to give ‘intuitive’ numerical values for non-Watson-Crick base pairs and associated steps. See the short communication titled Characterization of base pair geometry in the January 2016 issue of Computational Crystallography Newsletter (CCN).
The fiber program includes a new option, --pauling, for easy generation of Pauling & Corey triplex models of DNA/RNA with arbitrary base sequence. See my blogpost titled Pauling’s triplex model of nucleic acids is available in 3DNA.
Thomas Holder (PyMOL Principal Developer at Schrödinger, Inc.) has built a PyMOL wrapper to 3DNA fiber models. Now generating standard, regular DNA/RNA models in PyMOL is straightforward — thanks, Thomas!
DSSR (Dissecting the Spatial Structure of RNA)
Selected features of DSSR have been incorporated into Jmol (in collaboration with Robert Hanson, Jmol Principal Developer), and PyMOL (in collaboration with Thomas Holder). In Jmol application (via the Console window), one can now, for example, load =1ehz/dssr and then select hairpins; color red to see where the three hairpin loops are in 3D. The Jmol-DSSR web interface makes DSSR-enhanced visualization of nucleic acid structures in Jmol readily accessible to a broad user base, and has been employed in classes for educational purpose. A sample image of DSSR-derived cartoon-block representation via PyMOL is available for PDB entry 5dww, which has a G-quadruplex-duplex interface.
Since the publication of the Nucleic Acids Research paper in 2015, DSSR has been continuously refined and expanded, with a total of 36 new releases (from v1.2.8 to v1.6.4) as of this writing. Notably, the --json option provides DSSR-derived parameters in the simple, structured, and standard JSON format that can be easily parsed. This JSON output format is the (preferred) way for the outside world to interface with DSSR, and the Jmol-DSSR integration is built upon it. The --nmr option allows for batch processing of MODEL/ENDMDL-delineated NMR ensembles or trajectories of molecular dynamics (MD) simulations. Did you know that scripts and data files for reproducing the reported results are available in the DSSR-NAR paper section on the 3DNA Forum?
The User Manual is now 88-page long, covering nevertheless only the most common use cases of what DSSR has to offer. Miss a feature that you would like to have? Maybe it is already there or can be easily implemented in DSSR. Simply ask (on the 3DNA Forum), and I’ll try my best to help.
SNAP (Structures of Nucleic Acid-Protein complexes)
- SNAP aims to consolidate, refine, and significantly extend commonly used functionalities for DNA/RNA-protein structural analysis in one easy-to-use program. Currently in beta testing, SNAP is already fully functional, with features for characterizing the protein-nucleic acid interface and identifying amino acid-base pairing and stacking interactions.
A note for 3DNA/DSSR users in mainland China: It’s a pleasure to see the ~100 registrations on the 3DNA Forum with emails ending in .cn, 163.com, or qq.com etc., mostly from recent years. I’m planning a trip to China in 2017, and I’d be happy to meet some of you for academic exchanges and possible collaborations (学术交流、合作). If you’re interested, let’s get in touch!
Best regards,
Xiang-Jun
—
Dr. Xiang-Jun Lu (律祥俊)
Email: xiangjun@x3dna.org
Web: http://x3dna.org/
Forum: http://forum.x3dna.org/

In 1953, Pauling and Corey published an influential paper, titled A proposed structure for the nucleic acids, in Proc. Natl. Acad. Sci. (PNAS). Key features of the proposed model is summarized in their Letter to Nature, Structure of the Nucleic Acids, published in Nature on February 21, 1953.
We have formulated a structure for the nucleic acids which is compatible with the main features of the X-ray diagram and with the general principles of molecular structure, and which accounts satisfactorily for some of the chemical properties of the substances. The structure involves three intertwined helical polynucleotide chains. Each chain, which is formed by phosphate di-ester groups and linking β-D-ribofuranose or β-D-deoxyribofuranose residues with 3′, 5′ linkages, has approximately twenty-four nucleotide residues in seven turns of the helix. The helixes have the sense of a right-handed screw. The phosphate groups are closely packed about the axis of the molecule, with the pentose residues surrounding them, and the purine and pyrimidine groups projecting radially, their planes being approximately perpendicular to the molecular axis. The operation that converts one residue to the next residue in the polynucleotide chain is rotation by about 105° and translation by 3.4 Å.
This triplex model of nucleic acids, with phosphates in the center and bases on the outside, turned out to be fundamentally flawed. Yet, it played a significant role by prompting Watson and Crick in their discovery of the DNA double helix structure. While I’ve been aware of the Pauling triplex model from long ago, I had not read the original Pauling & Corey PNAS paper. Not surprisingly, I did not know what the triplex structure really looks like, other than some general ideas.
In a recent trip to Rutgers, Dr. Wilma Olson and I discussed the applications of fiber models collected in 3DNA. She drew my attention to the Pauling triplex model, and showed me Table 1 of the PNAS paper (see below), where the atomic coordinates for a nucleic acid repeating unit are listed.
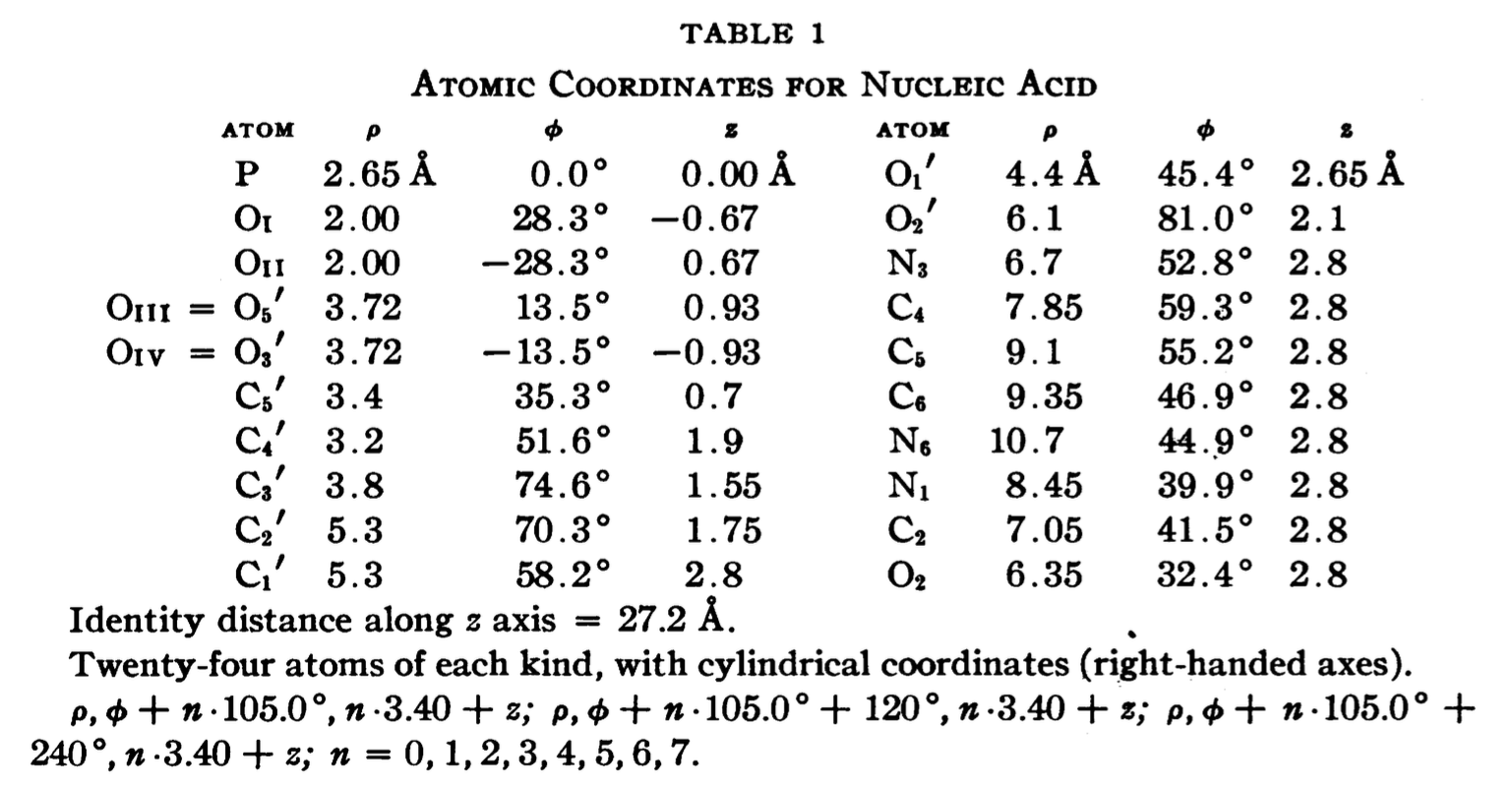
The cylindrical format is the same as that for the fiber models in 3DNA. It thus seems fitting to add this historically significant triplex model to the collection. Googling revealed many interesting historical notes and comments, e.g. The Pauling-Corey Structure of DNA, and a short video Linus Pauling’s triple DNA helix model, 3D animation with basic narration. However, I failed to find a program that I can use to generate such a triplex model with generic base sequence. I decided to add the fiber --pauling option so users can easily create such a triplex model in 3D, just as they do for a classic A- and B-DNA duplex. This process has turned out to be very educational (detailed below), and the end result should be of general interest.
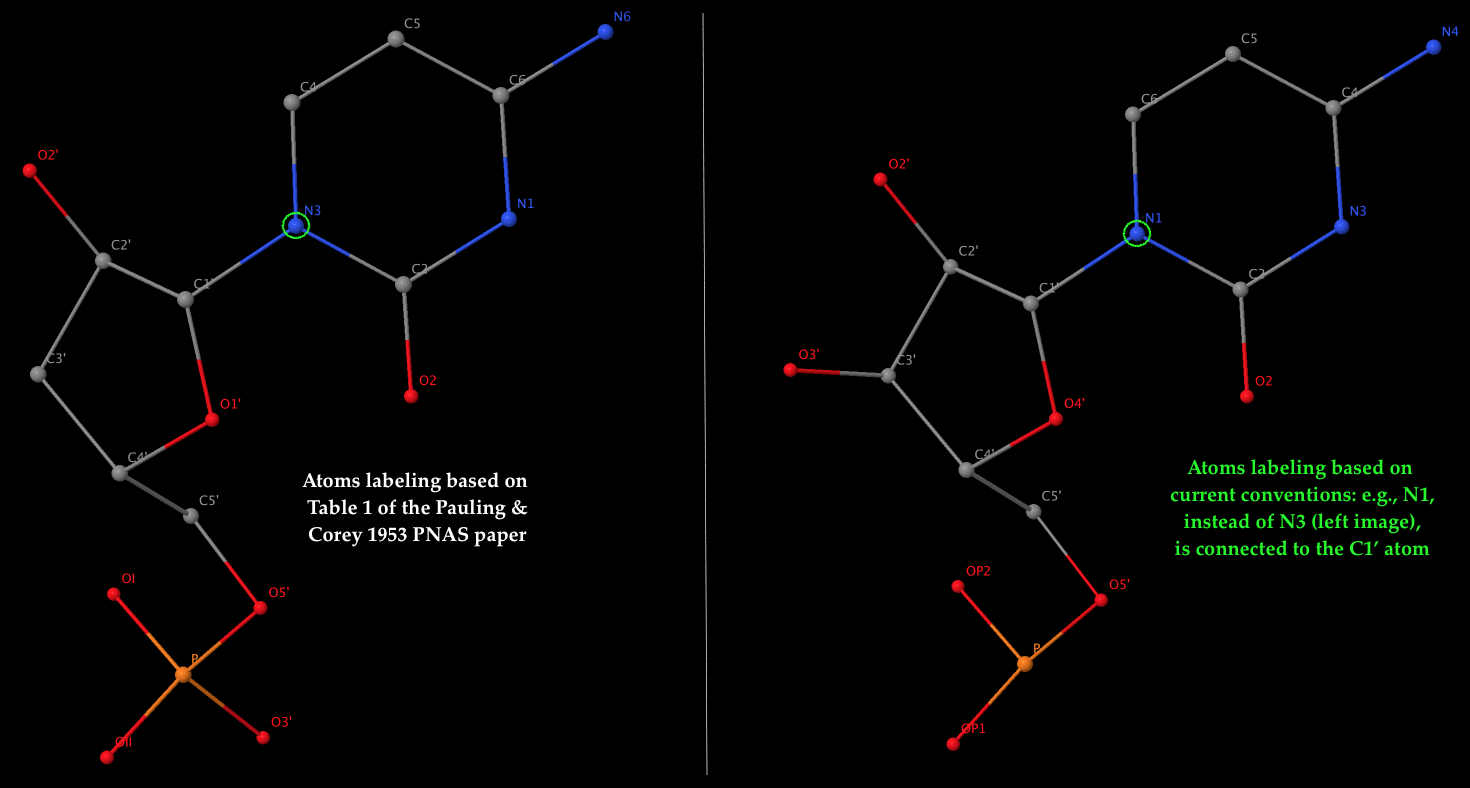
- The left 3D image shows the nomenclature of atoms used by Pauling & Corey (see Table 1 above), which is dramatically different from current conventions. As an example, it should be the N1 atom of cytosine (a pyrimidine base), not N3, that is connected to the sugar C1′ atom in nowadays nomenclature. The corrections apply not only to base atoms, but also to the sugar and phosphate groups. The revised atom labeling (as used in the PDB) is illustrated in the 3D image on the right.
- Table 1 corresponds to the ribose sugar since it contains an O2′ atom (see also the figure above). The triplex model constructed would be RNA, but can be ‘converted’ to DNA by simply removing the O2′ atom (see below).
- Only the atomic coordinates for cytosine are listed in Table 1. The 3DNA
mutate_bases program came handy to get the corresponding atomic coordinates for A, G, T, and U. This expansion allows for the generation of Pauling’s triplex models with an arbitrary combination of the five common bases (A, C, G, T, and U).
- With the new
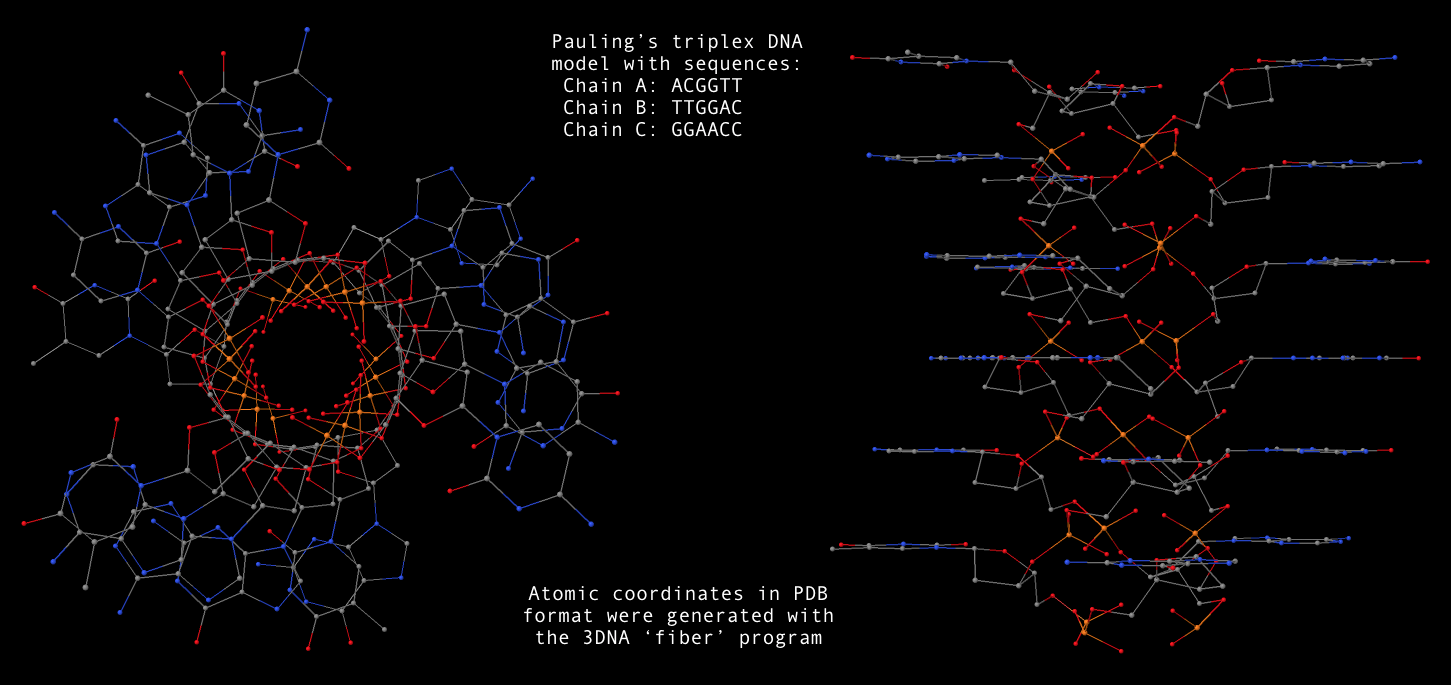
fiber --pauling option, now users can conveniently generate a Pauling’s triplex RNA/DNA model as shown below. Note that the one dash variant -pauling also works fine, with the additional -dna for DNA deoxyribose sugar. The PDB file (Pauling-triplex-mixed.pdb) with mixed DNA sequences can be downloaded, and the corresponding 3D image in top and side views is shown in the following figure.
fiber -pauling triplex-C10C10C10.pdb # default: 10 Cs per strand
fiber -pauling -seq=AAA triplex-A3A3A3.pdb # 3 As per strand
fiber -pauling -seq=AAAA:CCCC:GGGG Pauling-triplex-A4C4G4.pdb
fiber -pauling -seq=ACGGUU,UUGGAC,GGAACC Pauling-triplex-mixed.pdb
fiber --pauling-dna -seq=ACGGTT,TTGGAC,GGAACC Pauling-triplex-DNA.pdb
- With 3DNA’s
find_pair/analyze pair of programs, one can get the structural parameters corresponding to the Pauling triplex model. Not surprising, the repeating dinucleotide along each strand has a twist of 105°, and a rise of 3.4 Å. Notably, the sugar has a C2′-endo conformation.

3DNA contains 55 fiber models compiled from literature, plus a derived RNA model (as of v2.1). To the best of my knowledge, this is the most comprehensive collection of regular DNA/RNA models. Please see Table 4 of the 2003 3DNA NAR paper for detailed structural features of these models and references.
The 55 models are based on the following works:
- Chandrasekaran & Arnott (from #1 to #43) — the most well-known set of fiber models
- Alexeev et al. (#44-#45)
- van Dam & Levitt (#46-#47)
- Premilat & Albiser (#48-#55)
The utility program fiber makes the generation of all these fiber models in a simple, consistent interface, and produces coordinate files in either PDB or PDBML format. Of those models, some can be built with an arbitrary sequence of A, C, G and T (e.g., A-/B-/C-DNA from calf thymus), while others are of fixed sequences (e.g., Z-DNA with GC repeats). The sequence can be specified either from command-line or a plain text file, in either lower, UPPER, or MixED cases.
Once 3DNA in properly installed, the command-line interface is the most versatile and convenient way to generate, e.g., a regular double-stranded DNA (mostly, B-DNA) of arbitrary sequence. The command-help message (generated with fiber -h) is as below:
NAME
fiber - generate 55 fiber models based on Arnott and other's work
SYNOPSIS
fiber [OPTION] PDBFILE
DESCRIPTION
generate 55 fiber models based on the repeating unit from Arnott's
work, including the canonical A-, B-, C- and Z-DNA, triplex, etc
-xml output structure coordinates in PDBML format
-num a structure identification number in the range (1-55)
-m, -l brief description of the 55 fiber structures
-a, -1 A-DNA model (calf thymus)
-b, -4 B-DNA (calf thymus, default)
-c, -47 C-DNA (BII-type nucleotides)
-d, -48 D(A)-DNA ploy d(AT) : ploy d(AT) (right-handed)
-z, -15 Z-DNA poly d(GC) : poly d(GC)
-rna for RNA with arbitrary base sequence
-seq=string specifying an arbitrary base sequence
-single output a single-stranded structure
-h this help message (any non-recognized options will do)
INPUT
An structural identification number (symbol)
EXAMPLES
fiber fiber-BDNA.pdb
# fiber -4 fiber-BDNA.pdb
# fiber -b fiber-BDNA.pdb
fiber -a fiber-ADNA.pdb
fiber -seq=AAAGGUUU -rna fiber-RNA.pdb
fiber -seq=AAAGGUUU -rna -single fiber-ssRNA.pdb
OUTPUT
PDB file
SEE ALSO
analyze, anyhelix, find_pair
AUTHOR
3DNA v2.3-2016sept06, created and maintained by Xiang-Jun Lu (PhD)
Please post questions/comments on the 3DNA Forum: http://forum.x3dna.org/
Moreover, the w3DNA, 3D-DART web-interfaces, and the PyMOL wrapper make it easy to generate a regular DNA (or RNA) model, especially for occasional users or for educational purposes.
In principle, nothing is worth showing off with regard to 3DNA’s fiber model generation functionality. Nevertheless, this handy tool serves as a clear example of the differences between a “proof of concept” and a pragmatic software application. I initially decided to work on this tool simply for my own convenience. At that time, I had access to A-DNA and B-DNA fiber model generators, each as a separate program. Moreover, the constructed models did not comply to the PDB format in atom naming, among other subtitles.
I started with the Chandrasekaran & Arnott fiber models which I had a copy of data files. However, there were many details to work out, typos to correct, etc. to put them in a consistent framework. For other models, I had to read each original publication, and to type raw atomic cylindrical coordinates into computer. Again, quite a few inconsistencies popped up between the different publications with a time span over decades.
Overall, it was a quite tedious undertaking, requiring great attention to details. I am glad that I did that: I learned so much from the process, and more importantly, others can benefit from my effort. As I put in the 3DNA Nature Protocol paper (BOX 6 | FIBER-DIFFRACTION MODELS),
In preparing this set of fiber models, we have taken great care to ensure the accuracy and consistency of the models. For completeness and user verification, 3DNA includes, in addition to 3DNA-processed files, the original coordinates collected from the literature.
For those who want to understand what’s going on under the hood, there is no better way than to try to reproduce the process using, e.g., fiber B-DNA as an example.
From the very beginning, I had expected the 3DNA fiber functionality to serve as a handy tool for building a regular DNA duplex of chosen sequence. Over the years, the fiber program has gradually attracted attention from the community. The recent PyMOL wrapper by Thomas Holder is a clear sign of its increased popularity, and has prompted me to write this post, adapted largely from the one titled Fiber models in 3DNA make it easy to build regular DNA helices (dated Friday, October 9, 2009).
See also PyMOL wrapper to 3DNA fiber models

Given below is the content of the README file for fiber models in 3DNA:
1. The repeating units of each fiber structure are mostly based on the
work of Chandrasekaran & Arnott (from #1 to #43). More recent fiber
models are based on Alexeev et al. (#44-#45), van Dam & Levitt (#46
-#47) and Premilat & Albiser (#48-#55).
2. Clean up of each residue
a. currently ignore hydrogen atoms [can be easily added]
b. change ME/C7 group of thymine to C5M
c. re-assign O3' atom to be attached with C3'
d. change distance unit from nm to A [most of the entries]
e. re-ordering atoms according to the NDB convention
3. Fix up of problem structures.
a. str#8 has no N9 atom for guanine
b. str#10 is not available from the disk, manually input
c. str#14 C5M atom was named C5 for Thymine, resulting two C5 atoms
d. str#17 has wrong assignment of O3' atom on Guanine
e. str#33 has wrong C6 position in U3
f. str#37 to #str41 were typed in manually following Arnott's
new list as given in "Oxford Handbook of Nucleic Acid Structure"
edited by S. Neidle (Oxford Press, 1999)
g. str#38 coordinates for N6(A) and N3(T) are WRONG as given in the
original literature
h. str#39 and #40 have the same O3' coordinates for the 2nd strand
4. str#44 & 45 have fixed strand II residues (T)
5. str#46 & 47 have +z-axis upwards (based on BI.pdb & BII.pdb)
6. str#48 to 55 have +z-axis upwards
List of 55 fiber structures
id# Twist Rise Structure description
(dgrees) (A)
-------------------------------------------------------------------------------
1 32.7 2.548 A-DNA (calf thymus; generic sequence: A, C, G and T)
2 65.5 5.095 A-DNA poly d(ABr5U) : poly d(ABr5U)
3 0.0 28.030 A-DNA (calf thymus) poly d(A1T2C3G4G5A6A7T8G9G10T11) :
poly d(A1C2C3A4T5T6C7C8G9A10T11)
4 36.0 3.375 B-DNA (calf thymus; generic sequence: A, C, G and T)
5 72.0 6.720 B-DNA poly d(CG) : poly d(CG)
6 180.0 16.864 B-DNA (calf thymus) poly d(C1C2C3C4C5) : poly d(G6G7G8G9G10)
7 38.6 3.310 C-DNA (calf thymus; generic sequence: A, C, G and T)
8 40.0 3.312 C-DNA poly d(GGT) : poly d(ACC)
9 120.0 9.937 C-DNA poly d(G1G2T3) : poly d(A4C5C6)
10 80.0 6.467 C-DNA poly d(AG) : poly d(CT)
11 80.0 6.467 C-DNA poly d(A1G2) : poly d(C3T4)
12 45.0 3.013 D-DNA poly d(AAT) : poly d(ATT)
13 90.0 6.125 D-DNA poly d(CI) : poly d(CI)
14 -90.0 18.500 D-DNA poly d(A1T2A3T4A5T6) : poly d(A1T2A3T4A5T6)
15 -60.0 7.250 Z-DNA poly d(GC) : poly d(GC)
16 -51.4 7.571 Z-DNA poly d(As4T) : poly d(As4T)
17 0.0 10.200 L-DNA (calf thymus) poly d(GC) : poly d(GC)
18 36.0 3.230 B'-DNA alpha poly d(A) : poly d(T) (H-DNA)
19 36.0 3.233 B'-DNA beta2 poly d(A) : poly d(T) (H-DNA beta)
20 32.7 2.812 A-RNA poly (A) : poly (U)
21 30.0 3.000 A'-RNA poly (I) : poly (C)
22 32.7 2.560 Hybrid poly (A) : poly d(T)
23 32.0 2.780 Hybrid poly d(G) : poly (C)
24 36.0 3.130 Hybrid poly d(I) : poly (C)
25 32.7 3.060 Hybrid poly d(A) : poly (U)
26 36.0 3.010 10-fold poly (X) : poly (X)
27 32.7 2.518 11-fold poly (X) : poly (X)
28 32.7 2.596 Poly (s2U) : poly (s2U) (symmetric base-pair)
29 32.7 2.596 Poly (s2U) : poly (s2U) (asymmetric base-pair)
30 32.7 3.160 Poly d(C) : poly d(I) : poly d(C)
31 30.0 3.260 Poly d(T) : poly d(A) : poly d(T)
32 32.7 3.040 Poly (U) : poly (A) : poly(U) (11-fold)
33 30.0 3.040 Poly (U) : poly (A) : poly(U) (12-fold)
34 30.0 3.290 Poly (I) : poly (A) : poly(I)
35 31.3 3.410 Poly (I) : poly (I) : poly(I) : poly(I)
36 60.0 3.155 Poly (C) or poly (mC) or poly (eC)
37 36.0 3.200 B'-DNA beta2 Poly d(A) : poly d(U)
38 36.0 3.240 B'-DNA beta1 Poly d(A) : poly d(T)
39 72.0 6.480 B'-DNA beta2 Poly d(AI) : poly d(CT)
40 72.0 6.460 B'-DNA beta1 Poly d(AI) : poly d(CT)
41 144.0 13.540 B'-DNA Poly d(AATT) : poly d(AATT)
42 32.7 3.040 Poly(U) : poly d(A) : poly(U) [cf. #32]
43 36.0 3.200 Beta Poly d(A) : Poly d(U) [cf. #37]
44 36.0 3.233 Poly d(A) : poly d(T) (Ca salt)
45 36.0 3.233 Poly d(A) : poly d(T) (Na salt)
46 36.0 3.38 B-DNA (BI-type nucleotides; generic sequence: A, C, G and T)
47 40.0 3.32 C-DNA (BII-type nucleotides; generic sequence: A, C, G and T)
48 87.8 6.02 D(A)-DNA ploy d(AT) : ploy d(AT) (right-handed)
49 60.0 7.20 S-DNA ploy d(CG) : poly d(CG) (C_BG_A, right-handed)
50 60.0 7.20 S-DNA ploy d(GC) : poly d(GC) (C_AG_B, right-handed)
51 31.6 3.22 B*-DNA poly d(A) : poly d(T)
52 90.0 6.06 D(B)-DNA poly d(AT) : poly d(AT) [cf. #48]
53 -38.7 3.29 C-DNA (generic sequence: A, C, G and T) (depreciated)
54 32.73 2.56 A-DNA (generic sequence: A, C, G and T) [cf. #1]
55 36.0 3.39 B-DNA (generic sequence: A, C, G and T) [cf. #4]
-------------------------------------------------------------------------------
List 1-41 based on Struther Arnott: ``Polynucleotide secondary structures:
an historical perspective'', pp. 1-38 in ``Oxford Handbook of Nucleic
Acid Structure'' edited by Stephen Neidle (Oxford Press, 1999).
#42 and #43 are from Chandrasekaran & Arnott: "The Structures of DNA
and RNA Helices in Oriented Fibers", pp 31-170 in "Landolt-Bornstein
Numerical Data and Functional Relationships in Science and Technology"
edited by W. Saenger (Springer-Verlag, 1990).
#44-#45 based on Alexeev et al., ``The structure of poly(dA) . poly(dT)
as revealed by an X-ray fiber diffraction''. J. Biomol. Str. Dyn, 4,
pp. 989-1011, 1987.
#46-#47 based on van Dam & Levitt, ``BII nucleotides in the B and C forms
of natural-sequence polymeric DNA: a new model for the C form of DNA''.
J. Mol. Biol., 304, pp. 541-561, 2000.
#48-#55 based on Premilat & Albiser, ``A new D-DNA form of poly(dA-dT) .
poly(dA-dT): an A-DNA type structure with reversed Hoogsteen Pairing''.
Eur. Biophys. J., 30, pp. 404-410, 2001 (and several other publications).

Recently, I heard from Thomas Holder, the PyMOL Principal Developer (Schrödinger, Inc.), that he had written a wrapper to the 3DNA fiber command. This PyMOL wrapper is implemented as part of his versatile PSICO library (see the PyMOL Wiki page Psico for details), and exposes the 55 fiber models based on Arnott and other’s work to the wide PyMOL user community. Moreover, the wrapper can be accessed directly from PyMOL (without installing PSICO), as shown below with an example:
PyMOL> run https://raw.githubusercontent.com/speleo3/pymol-psico/master/psico/creating.py
PyMOL> fiber CTAGCG
The resulting fiber model is the default B-form DNA of calf thymus, with twist of 36.0° and rise of 3.375 Å (see figure below). Note that cases in base sequence do not matter, so fiber ctagcg or fiber CTAgcg will give the same result.
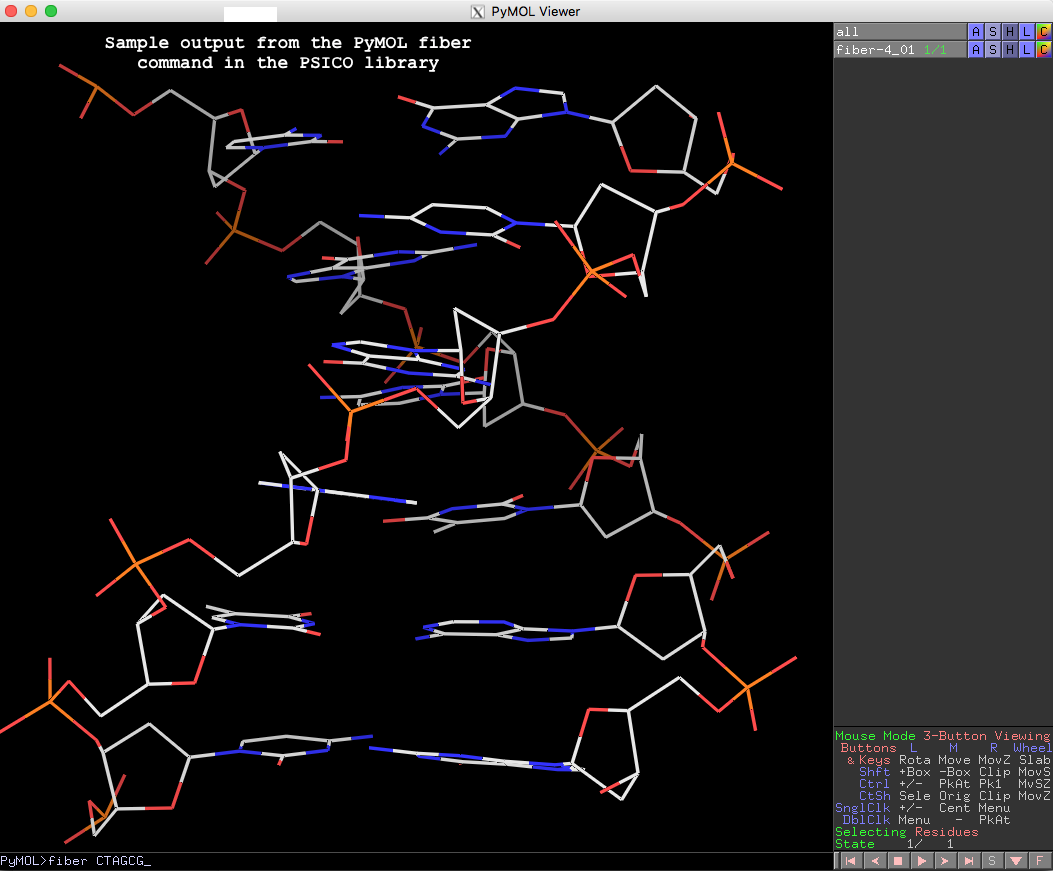
Running PyMOL>help fiber gives the following detailed usages info, which should be sufficient to get one started with this fiber tool in PyMOL.
PyMOL> help fiber
DESCRIPTION
Run X3DNA's "fiber" tool.
For the list of structure identification numbers, see for example:
http://xiang-jun.blogspot.com/2009/10/fiber-models-in-3dna.html
USAGE
fiber seq [, num [, name [, rna [, single ]]]]
ARGUMENTS
seq = str: single letter code sequence or number of repeats for
repeat models.
num = int: structure identification number {default: 4}
name = str: name of object to create {default: random unused name}
rna = 0/1: 0=DNA, 1=RNA {default: 0}
single = 0/1: 0=double stranded, 1=single stranded {default: 0}
EXAMPLES
# environment (this could go into ~/.pymolrc or ~/.bashrc)
os.environ["X3DNA"] = "/opt/x3dna-v2.3"
# B or A DNA from sequence
fiber CTAGCG
fiber CTAGCG, 1, ADNA
# double or single stranded RNA from sequence
fiber AAAGGU, name=dsRNA, rna=1
fiber AAAGGU, name=ssRNA, rna=1, single=1
# poly-GC Z-DNA repeat model with 10 repeats
fiber 10, 15
Thanks to Thomas, for making another connection between PyMOL and 3DNA/DSSR. The other one is the DSSR-plugin for PyMOL to create “block” shaped cartoons for nucleic acid bases and base pairs.
See also 3DNA fiber models

As of release v2.3-2016sept06, the C source code of the 3DNA software package is available. The code can be found in the $X3DNA/src folder of the distributed tarballs for Linux, Mac OS X, and Windows. Since 3DNA is written in pure ANSI C, it can be compiled without changes on any platform with a modern C compiler.
The original codebase of 3DNA was written around year 2000. Up until v2.3, the infrastructure of 3DNA has remained stable for 16 years. During the time, 3DNA has been widely adopted in other bioinformatics pipelines and cited over 1,500 times. Over the years, I’ve received quite a few requests for 3DNA source code. However, due to complications of various factors (including software licensing), 3DNA had only been distributed in executable forms for the crucial C programs. Now, the C code of 3DNA is finally open source!
As before, users need to register on the 3DNA Forum to download the software. The download page also includes x3dna-v2.0.tar.gz that accompanied the 2008 Nature Protocols paper, and x3dna-v1.5.tar.gz that corresponded to the 2003 Nucleic Acids Research paper. Other than minor revisions to pass strict gcc compiler options, the v1.5 and v2.0 codebases are kept as they were. 3DNA is backward-compatible as far as the key base-pair parameters are concerned. Moreover, between v1.5 and v2.0, the command-line interface stays the same. The two previous versions are released for historical reasons. For example, one may notice some obvious “similarities” between 3DNA v1.5 and RNAView.
The development of DSSR and SNAP will push 3DNA into a brand new version (v3), which contains significant changes in functionality and interface, and is no longer compatible with previous versions. I intend to keep 3DNA v2.3 in a ‘maintenance’ mode: no new features are planed, but bug reports and user questions will be promptly addressed on the 3DNA Forum, as always. Making 3DNA open source should help further prompt its adoptions, and adaptations in structural bioinformatics of nucleic acids.
There are numerous types of software licenses, but none of them seems to be a good fit for my purpose. As a result, I’ve come up with a permissive “citation-ware” license with contents as below:
3DNA is a suite of software programs for the analysis,
rebuilding and visualization of 3-Dimensional Nucleic Acid
structures. Permission to use, copy, modify, and distribute
this suite for any purpose, with or without fee, is hereby
granted, and subject to the following conditions:
At least one of the 3DNA papers must be cited, including the
following two primary ones:
1. Lu, X. J., & Olson, W. K. (2003). "3DNA: a software
package for the analysis, rebuilding and visualization
of three‐dimensional nucleic acid structures." Nucleic
Acids Research, 31(17), 5108-5121.
2. Lu, X. J., & Olson, W. K. (2008). "3DNA: a versatile,
integrated software system for the analysis,
rebuilding and visualization of three-dimensional
nucleic-acid structures." Nature Protocols, 3(7),
1213-1227.
THE 3DNA SOFTWARE IS PROVIDED "AS IS", WITHOUT EXPRESSED OR
IMPLIED WARRANTY OF ANY KIND.
Any 3DNA-related questions, comments, and suggestions are
welcome and should be directed to the open 3DNA Forum
(http://forum.x3dna.org/).

Upon user requests, I’ve recently introduced the --block-color option to DSSR, available as of v1.5.2-2016apr02. As its name implies, the --block-color option facilitate user customization of PyMOL rendered colors of the base rectangular blocks or their edges (e.g., the minor-groove) directly from the command-line. A simple example goes like this: --block-color='A blue; T red', which makes A colored blue and T colored red. As detailed below, the new option is very flexible with regard to the specification of colors, bases, or some edges to highlight. Before that, a little background is in order.
Background info
The DSSR cartoon-block representation follows the color convention of the original 3DNA blocview script, where A is red; C is yellow; G is green; T is blue; and U is cyan. If I remember correctly, the blocview coloring was based on the scheme adopted by the Nucleic Acid Database (NDB). To allow for some flexibility, 3DNA includes a config file named $X3DNA/config/raster3d.par where users can change the RGB values of the corresponding bases. However, I do not know if any user has ever bothered to play around with the configuration file for customized base colors.
Over the years, blocview-generated images have become popular, due to its simplicity, and (maybe more importantly) its endorsement by the NDB and PDB for nucleic acid structures. Via NDB, the blocview-generated images have also been used in RNA FRABASE 2.0 and RNA Structure Atlas. Nevertheless, the blocview script has several dependencies: MolScript for protein or DNA/RNA backbone ribbons, render from Raster3D for rendering, and ImageMagick for image processing. Moreover, the blocview script used by NDB/PDB is (likely to be) based on 3DNA v1.5, the last version before I left Rutgers in 2002.
Over the years, 3DNA has been continuously refined, with significant changes introduced in v2.0 around 2008 to accompany the Nature Protocols paper. Currently at v2.3, the codebase for 3DNA version 2 is in maintenance mode: the software will still be supported with identified bugs fixed, but no more new feature is planned. 3DNA version 3, as represented by DSSR and SNAP, is the way to go.
DSSR has no third-party dependencies
While creating DSSR, I set it as one of the design goals to make the program fully self-contained, without any third-party dependencies. Connections to other tools are clearly delineated via text files. If anything goes wrong, one can easily identify where the problem is. Experience over the past few years has unambiguously proved the effectiveness of this zero-dependency approach. Other than being directly distributed with an operating system, DSSR is the easiest to get up and running. Moreover, DSSR can be easily integrated into other pipelines, including Jmol and PyMOL, among many other bioinformatics tools.
For the cartoon-block representation, DSSR produces .r3d files that can be loaded into PyMOL, mixed and matched with other visualization styles PyMOL has to offer. No more direct dependencies on MolScript, Raster3D, and ImageMagick as is the case for blocview. It is also worth mentioning that DSSR does not need PyMOL to run. DSSR and PyMOL are connected via .r3d files, a process which can be streamlined with the Dssr_block PyMOL plugin.
DSSR releases before v1.5.2-2016apr02 have the color coding of base blocks fixed within the source code, following the default style of blocview. Over the past few months, I’ve received at least two explicit requests on customizing the default colors of DSSR-generated base blocks. The --block-color option has been introduced for this purpose.
Details of the --block-color option
The general format of the option is as follows:
--block-color='id color [; id2 color2 ...]'
id can be A, C, G, T, U, or the degenerated IUPAC code, including R, Y, N etc. See UPAC nucleotide code for details.
id can also be minor, major, upper, bottom, wc-edge to specify one of the six faces of a 3D rectangular block. See Fig.1D of the DSSR paper for details.

id can further be GC, AT, GU, pair, and variants thereof, to specify the colors of the corresponding long base-pair rectangular blocks.
color can be a common name (144 total), as specified in the RGB Color website. For example, red, magenta, light gray etc.
color can also be a single number in the range [0, 1] or [0, 255] to specify a shade of gray. DSSR repeat the number twice to get the RGB triple consisting of the same number.
color can further be a set of three space-delimited numbers to specify the RGB triple. Again, the number can be in [0, 1] or [0, 255]. Moreover, the three numbers can be put in square brackets. For example --block-color='A 0 1 1' and --block-color='A [0 1 1]' specify adenine to be colored with RGB triple [0 1 1] (aqua/cyan, corresponding to --block-color='A cyan').
- More than one identity (bases) can be specified, separated by
; (,, :, or | also works). Note: within the PyMOL dssr_block plugin, only | or : can be used as a separator: comma (,) or semicolon (;) cannot be used as a separator within a PyMOL command argument (thanks to Thomas Holder for drawing this point to my attention).
- Case does not matter when specifying
id or color. So either ‘A’ or ‘a’, and ‘blue’ or ‘Blue’ or ‘BLUE’ can be used to make adenine blue: --block-color='a blue'.
Some example usages
While the above description may appears to be quite complicated, the actual usage of the --block-color option is very straightforward. As always, the cases are best made with concrete examples, as shown below using the classic Dickerson B-DNA dodecamer 355d.
# all bases in blue
x3dna-dssr -i=355d.pdb --cartoon-block=orient --block-color='N blue' -o=355d-all-blue.pml
#
# all WC-pairs in red, with the minor-groove edge in 'dim gary'
x3dna-dssr -i=355d.pdb --cartoon-block=orient --block-color='wc-pair red; minor dim gray' -o=355d-pair-minor.pml
#
# thymine (T) in purple, and the upper (+z) face in white
# see Figure below, which shows the two bases in WC-pairs are anti-parallel
x3dna-dssr -i=355d.pdb --cartoon-block=orient --block-color='T purple; upper 1' -o=355d-T-upper.pml
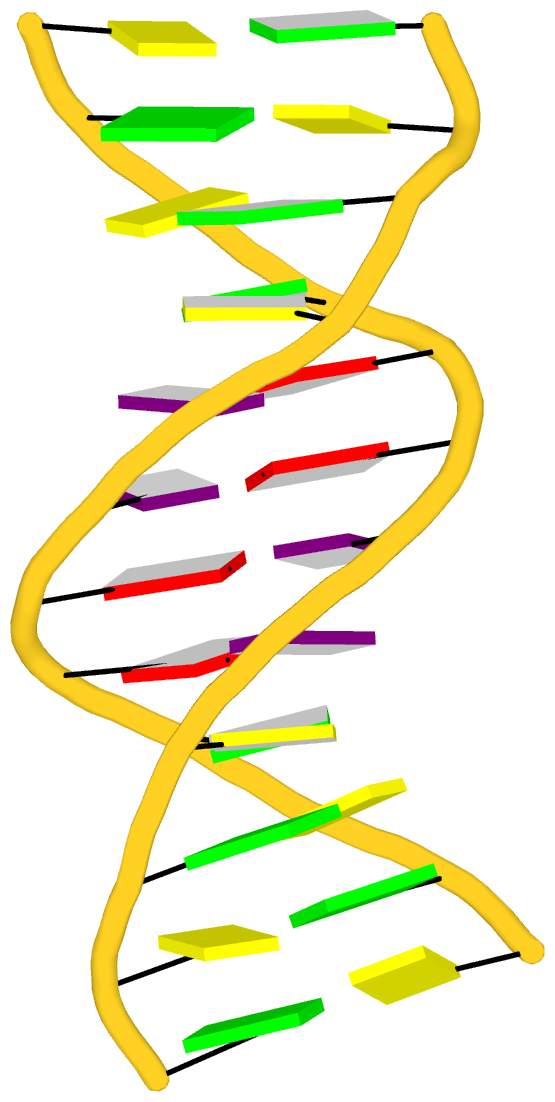

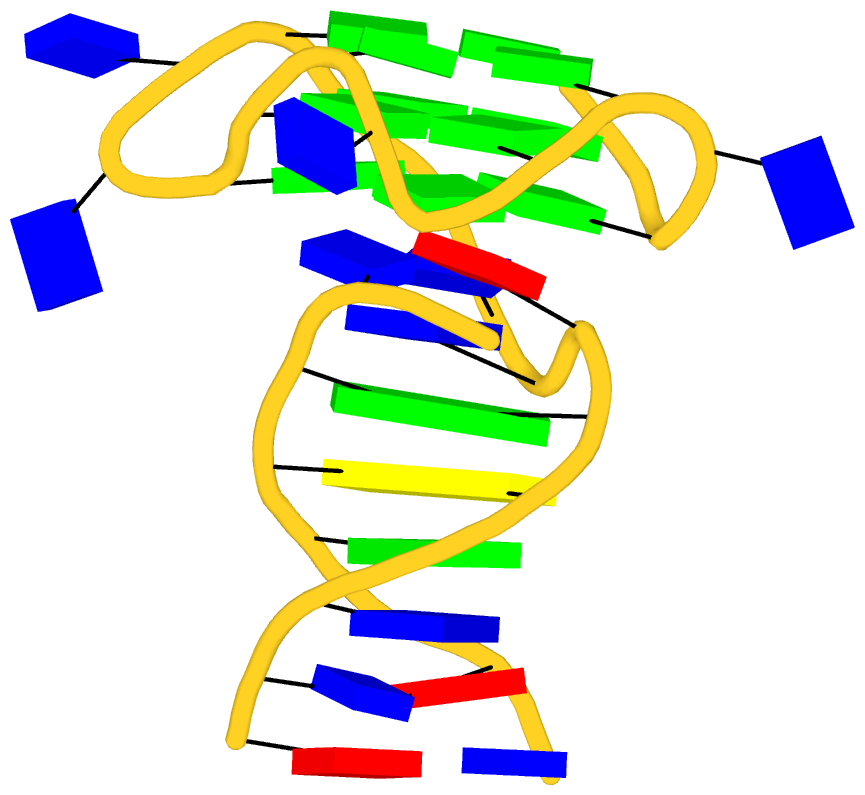
Recently I read the article titled Structural Insights into the Quadruplex−Duplex 3′ Interface Formed from a Telomeric Repeat: A Potential Molecular Target by Krauss et al.. I quickly ran DSSR on the corresponding PDB entry is 5dww. Not surprisingly, DSSR can automatically identify reported key structural features (see output file 5dww.out for details), including the TAT triplet at the quadruplex−duplex junction, and the three G-quartets. Note that the result is based on biological assembly 1 in PDB file 5dww.pdb1 since the asymmetric unit contains four such molecules.
List of 4 multiplets
1 nts=3 TAT 1:A.DT17,1:A.DA19,1:B.DT7
2 nts=4 GGGG 1:A.DG1,1:A.DG5,1:A.DG9,1:A.DG14
3 nts=4 GGGG 1:A.DG2,1:A.DG6,1:A.DG10,1:A.DG15
4 nts=4 GGGG 1:A.DG3,1:A.DG7,1:A.DG11,1:A.DG16
As its title suggests, however, this blog post is about the cartoon-block representations. Four styles of such schematics are shown below, which can all be easily generated using DSSR/PyMOL.
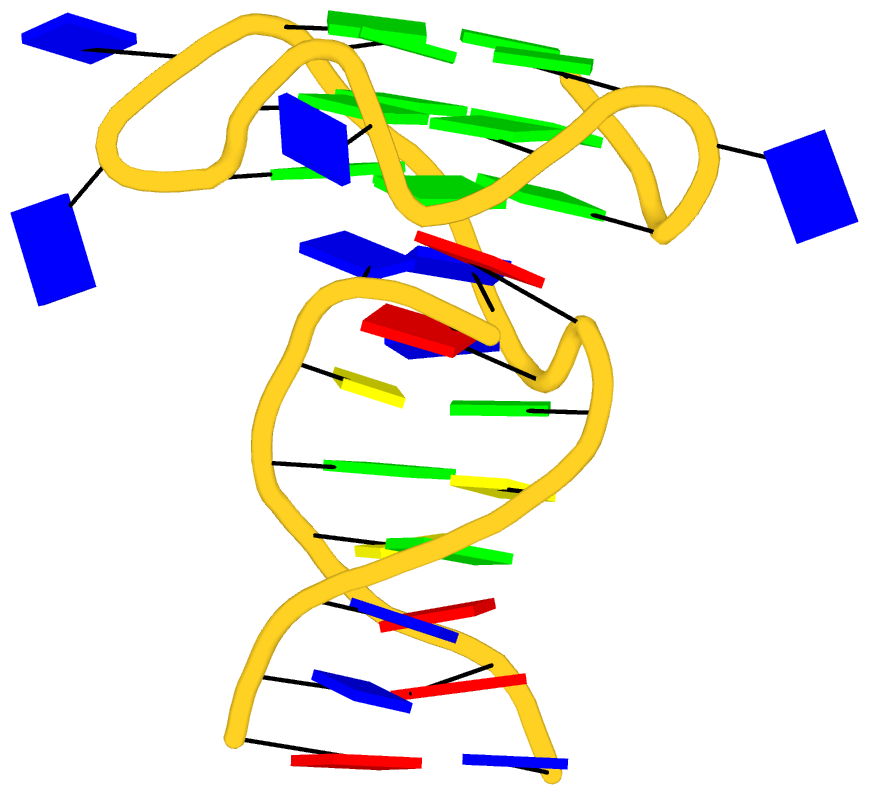 |
 |
| in default style |
with base-pair blocks |
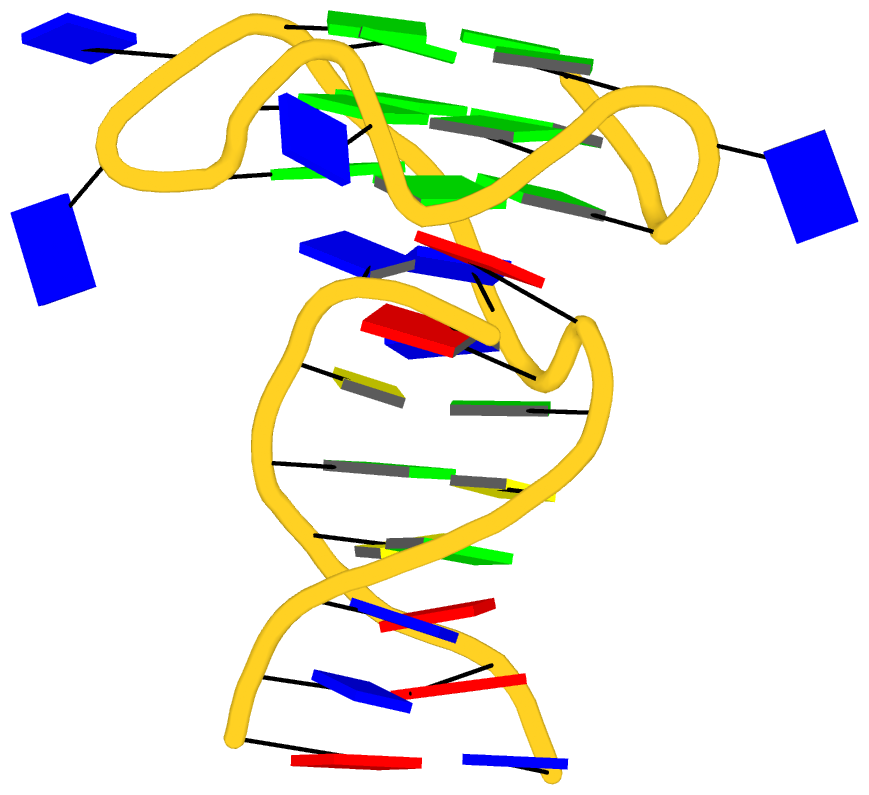 |
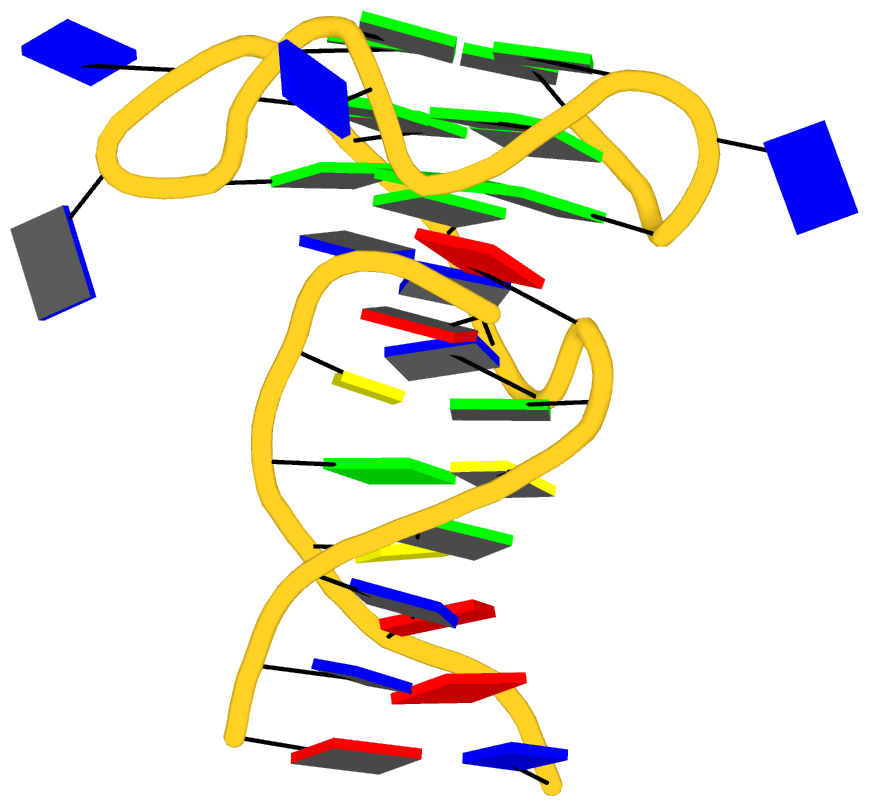 |
| minor-groove highlighted |
top-face highlighted |
The cartoon-block representations possess unique features not seen elsewhere. With the help of the dssr_block in PyMOL, they are extremely easy to generate. Such schematics are likely to become popular in illustrations of nucleic acid structures.

Over the past couple of years, one of the most significant achievements of DSSR has been its integration into Jmol and PyMOL, two widely used molecular graphics programs. None of the projects had been ‘planned’, and I am honored to have the opportunities collaborating directly with Bob Hanson (Jmol) and Thomas Holder (PyMOL). The integrations make salient features of DSSR readily accessible to the Jmol and PyMOL user communities. Moreover, Jmol and PyMOL take different approaches to interoperate with DSSR, and so far they have employed separate features that the program has to offer.
Key features of DSSR
DSSR was implemented in strict ANSI C as a self-contained command-line program. The binaries for common operating systems (Mac OS X, Linux and Windows) are tiny (<1MB), and without runtime dependencies on third-party libraries. DSSR also comes with an extensive PDF user manual.
Since its initial release in early 2013, DSSR has been continuously refined/expanded based on user feedback and my improved knowledge of RNA structures. User questions are always promptly addressed on the public 3DNA Forum. Over the years, DSSR has gradually established itself as an accountable software product.
The small size, zero configuration, extensive features, and robust performance make DSSR ideal to be integrated into other bioinformatics tools.
DSSR and Jmol
From the very beginning, Jmol has been employing a web-service at Columbia University, where all DSSR analyses take place. In addition to the sample DSSR-Jmol web interface, DSSR is also directly accessible from the console (see Fig.1 below). Jmol includes a sophisticated SQL syntax to drill down the various DSSR-derived structure features. Search ‘DSSR’ on the Jmol/JSmol interactive scripting documentation for details.
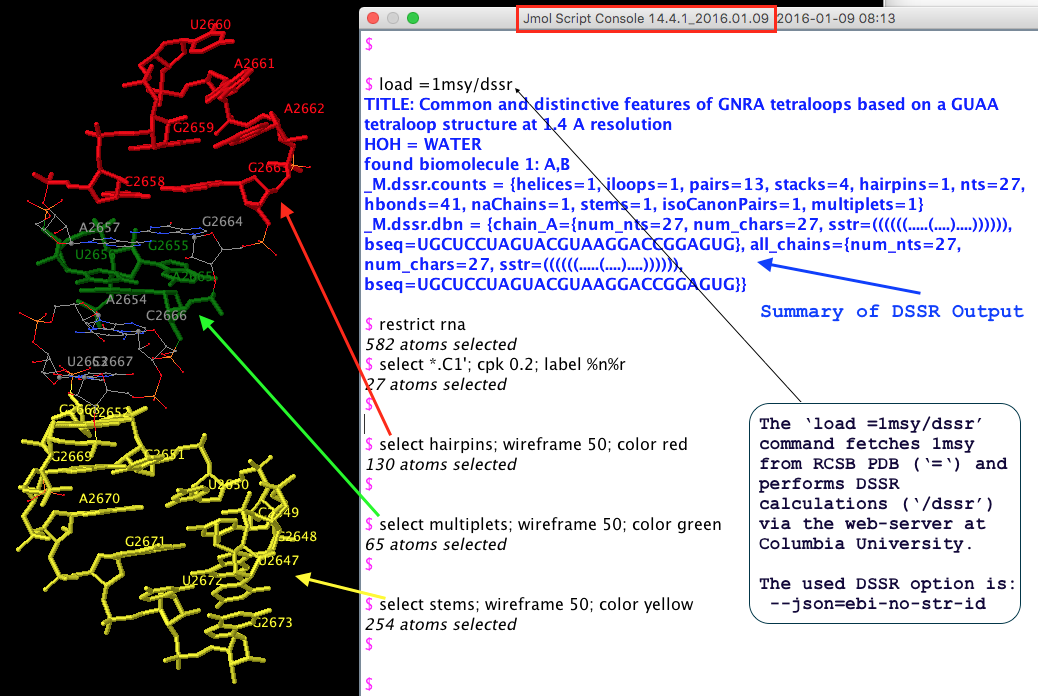 Fig. 1 DSSR is available from the Jmol/JSmol console via scripting.
Fig. 1 DSSR is available from the Jmol/JSmol console via scripting.
The initial version of the integration (Jmol v14.2) was facilitated by the DSSR --jmol option to produce a Jmol-specific (e.g., residue id [C]2658:A) plain text output. However, ad hoc text file are rigid and fragile for programs to communicate with. As DSSR had been evolving, changes to existing features or newly added functionality were known to break the established DSSR-Jmol interface. Having to write extra code to maintain the same old --jmol output did not feel right.
JSON (JavaScript Object Notation) came to the rescue! The current DSSR-Jmol integration (Jmol v14.4) takes advantage of JSON, a standard, lightweight data-interchange format. Since JSON is structured, parsing its contents is straightforward. DSSR and Jmol can evolve independently, as always, but they no longer need to worry about touching each other’s toes.
Overall, Jmol has incorporated the most fundamental analysis features of DSSR. The Jmol SQL mini-language is very powerful for selecting arbitrary DSSR parameters. Background information about this collaboration can be found in the blog post Jmol and DSSR.
DSSR and PyMOL
So far, the DSSR-PyMOL integration has focused on visualization, i.e., the cartoon-block schematic representations of DNA/RNA structures. Moreover, instead of relying on a remote DSSR web-service as for Jmol, the PyMOL dssr_block command calls a locally installed DSSR executable for the job. As illustrated in the blogpost DSSR base blocks in PyMOL, interactively, the ‘dssr_block’ command makes it trivial to incorporate the highly effective rectangular blocks into PyMOL.
From early on, 3DNA includes the blocview script (first written in Perl, later converted to Ruby) to generate schematic images in the ‘best view’, by combining block representation of bases with backbone ribbon of proteins or nucleic acids. The script is essentially a glue, calling MolScript, Raster3D, ImageMagick, and several 3DNA utility programs to perform various tasks. With these dependencies, it’s a bit involved to set up blocview. Nevertheless, the resultant images are simple and revealing, and are still being used by NDB and RCSB PDB (among others) as of today.
DSSR does not depend on MolScript and Raster3D, or any other programs to generate .r3d output of rectangular blocks. The schematic blocks can be directly fed into PyMOL, combined with other representations, and ray-traced for high resolution images. The integration of DSSR into PyMOL by the dssr_block command is likely to prompt an even wider adoption of the cartoon-block representation. In this regard, it is well worth noting the news item “dssr_block is a wrapper for DSSR (3dna) and creates block-shaped nucleic acid cartoons” on the main page of PyMOLWiki (see Fig. 2 below). It will certainly bring this neat feature into the attention of many PyMOL users.
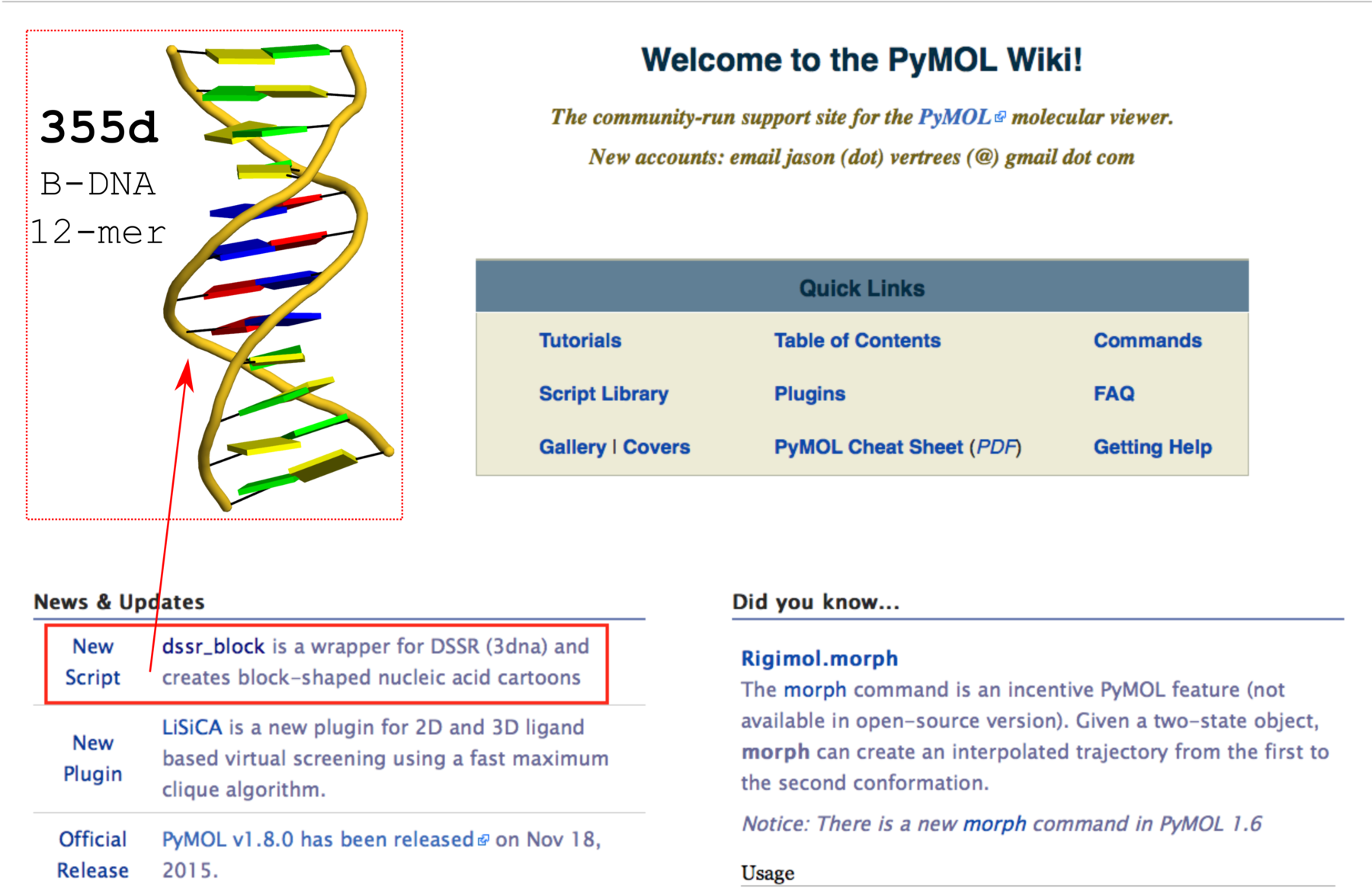
Fig. 2 Screenshot of the PyMOLWiki main page (2016-01-27) with ‘dssr_block’ in the news. A sample cartoon-block image of 355d is inserted as an example.
Integration of DSSR analysis results into PyMOL is underway, using the same JSON output. Before long, PyMOL users should be able to have access to the numerous DNA/RNA structural features derived by DSSR as in Jmol, along with the cartoon-block images enabled by dssr_block. Background information about DSSR-PyMOL can be found in blog post Open invitation on writing a DSSR plugin for PyMOL.
Notes
- The DSSR-Jmol and DSSR-PyMOL integrations are two salient examples of what can be achieved via direct collaboration of dedicated scientists with complementary expertise. In addition to benefit the involved projects in particular and the (structural biology) community at large, technical and scientific advances are more likely to be achieved.
- Both projects are still on going, with continued refinements of existing functionality and additions of new features. As an example, it is desirable and likely that Jmol would allow local access to DSSR for efficiency and data privacy.
- JSON is the way to go for connecting DSSR to the outside world. Period. The obsolete
--jmol will be removed from the next release of DSSR (v1.5). The default plain text output is useful for easy comprehension and will stilled be maintained. But do not count on its exact format for computer parsing — occasional changes to existing items are likely, and new features are bound to be added.
- If you’d like to incorporate DSSR into your pipeline and need some customizations of its output, please let me know. It’s always easier to set things right at the source than to fix them downstream. Where practical, I’ll try to implement your requested features, quickly. Working together, we can and will build a better world.

















