The blocview script in 3DNA has been created as a handy tool to effectively reveal key features of small to medium-sized nucleic acid structures. Specifically, the bloc part of the name means ‘block’, i.e., the rectangular block in Calladine-Drew style schematic representation to distinguish bases by size (larger purine vs. smaller pyrimidine), identity (red for A, yellow for C, green for G, and blue for T), and groove (minor edge in black). The view part stands for the most extended view, as defined by the principal axes of inertia. Implementation-wise, blocview calls several 3DNA utility programs and MolScript (for protein ribbons and nucleic acid backbone rods) to prepare the scenes, and then uses Raster3D (specifically, render) or PyMol to generate a PNG image.
The blocview script was originally written in Perl. As of 3DNA v2.1, I decided to switch the scripting language to Ruby for its consistent object-oriented style, succinct and flexible syntax. Previously available Perl scripts are now moved out of the default 3DNA executable directory $X3DNA/bin/ into $X3DNA/perl_scripts/. The blocview script has been re-written in Ruby and set as the default (at $X3DNA/bin/blocview); the original Perl version is renamed blocview.pl (at $X3DNA/perl_scripts/blocview.pl) to avoid confusion. The command line help message, available via blocview -h, is as below:
------------------------------------------------------------------------
Generate a schematic image which combines base block representation
with protein ribbon. The image has informative color coding for the
nucleic acid part and is set in the "best view" by default. Raster3D
(or PyMOL) and ImageMagick must be installed.
Usage:
blocview [options] PDBFile
Examples:
blocview -i 355d.png 355d.pdb
# generate image '355d.png'; display 355d.png
Options:
------------------------------------------------------------------------
--imgfile, -i <s>: name of image file (default: blocview.png)
--r3dfile, -r <s>: name of .r3d file (default: blocview.r3d)
--dpi-pymol, -d <i>: create PyMOL ray-traced image at specific DPI
--scale, -s <f>: set scale factor (for 'render' of Raster3D)
--xrot, -x <f>: rotation angle about x-axis
--yrot, -y <f>: rotation angle about y-axis
--zrot, -z <f>: rotation angle about z-axis
--original, -o: use original coordinates
--ball-and-stick, -b: get a ball-and-stick image
--p-base-ring, -c: use only P and base ring atoms
--no-ds, -n: do not show double-helix ribbon
--protein, -p: set best view based on protein atoms
--all, -a: set best view based on all atoms
--version, -v: Print version and exit
--help, -h: Show this message
Using the x-ray crystal structure of d(GGCCAATTGG) complexed with netropsin (1z8v) in the minor groove as an example, the command to run is as follows:
blocview -i 1z8v.png 1z8v.pdb # The following two forms are also fine # blocview --imgfile 1z8v.png 1z8v.pdb # blocview --imgfile=1z8v.png 1z8v.pdb # The Perl version can be run like this: # $X3DNA/perl_scripts/blocview.pl -i=1z8v.png 1z8v.pdb
The image, named 1z8v.png, is shown below. Note that it is generated automatically from the PDB-formatted data file 1z8v.pdb. In this representation, one can see clearly that there are two unpaired Gs (green block) at each 5′-end of the two DNA chains (red and yellow rods), and a drug molecule (ball-and-stick) binds in the minor groove (black edge of the rectangular blocks). Moreover, the deformation in propeller and buckle is obvious in this schematic presentation.
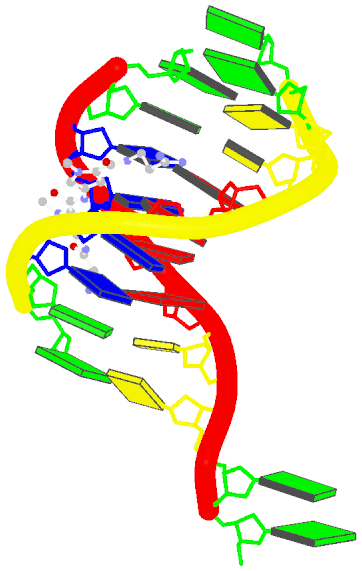
Over the years, blocview-generated images have been used in NDB for virtually all nucleic acid structures (see for example, the NDB atlas gallery for x-ray drug-DNA complexes). It’s worth noting that such simple images have also be adopted by the RCSB PDB, prominently at the summary page, for nucleic acid containing structures (see PDB entry 1z8v). Given the effectiveness of blocview-generated schematic representation and its adoption by the NDB and PDB, I’m hopeful that blocview will be more widely used by the general DNA/RNA structure community. As always, I value user’s feedback in continuously refining the script.
