 March 2026
March 2026
 February 2026
February 2026
Cover image provided by X3DNA-DSSR, an NIGMS National Resource for Structural Bioinformatics of Nucleic Acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).
As the developer of DSSR, I am thrilled to see its application in cutting-edge research across multiple disciplines. Below is a list of four recent publications that highlight how DSSR has been utilized, underscoring its versatility and significance in structural bioinformatics.
In the Geng et al. (2025) Nucleic Acids Research (NAR) paper, titled 'Revealing hidden protonated conformational states in RNA dynamic ensembles', DSSR is simply cited as follows:
All bp geometries, hydrogen-bond, backbone, stacking, and sugar dihedral angles were calculated using X3DNA-DSSR [77].
In the preprint by Gordan et al. (2025), titled 'High-throughput characterization of transcription factors that modulate UV damage formation and repair at single-nucleotide resolution', DSSR is cited as follows:
Step base stacking, base pair shift, base pair slide, interbase angle, pseudorotation angle, and sugar puckering classifications of nucleobases were computed using X3DNA-DSSR (v2.5.0)75. Base stacking was defined as the overlapping polygon area in Å2 when projecting the dipyrimidine base ring atoms (excluding exocyclic atoms) into the mean base pair plane76. The sugar ring pseudorotation phase angle of each pyrimidine was also calculated using X3DNA-DSSR as described by Altona, C. & Sundaralingam, M.77 Interbase angle was defined as sqrt(propeller2+buckle2) per the X3DNA-DSSR documentation.
Figure 6: TF Binding Induces Structural Distortion Favorable to UV Dimerization is highly informative, particularly panel (a), which illustrates the ensemble of structural parameters that predispose dipyrimidines to cyclobutane pyrimidine dimers (CPD) or 6-4 pyrimidine-pyrimidones (6-4 PP) formation. DSSR is designed as an integrated software tool, offering a comprehensive suite of structural parameters not found in any other single tool I am aware of. Despite this, the innovative use of DSSR by Gordan et al. exceeds my expectations and demonstrates its versatility.
In the preprint by Kubaney et al. (2025) from the Baker group, titled 'RNA sequence design and protein-DNA specificity prediction with NA-MPNN', DSSR is cited as follows:
On the pseudoknot subset, we evaluate additional structure‐ and reactivity‐based metrics. DSSR v2.3.241 is used to extract the ground‐truth secondary structure from the native crystal structures. For each designed sequence, RibonanzaNet predicts 2A3 reactivity profiles, from which we compute predicted OpenKnot scores (see https://github.com/eternagame/OpenKnotScore)31 using the predicted reactivity together with the DSSR ground truth.
In a recent NSMB paper from the Baker group, titled 'Computational design of sequence-specific DNA-binding proteins', 3DNA is cited as follows:
RIF docking of scaffolds onto DNA targets (DBP design step 1) Structures of B-DNA for each target (Supplementary Table 2) were generated by (1) using the DNA portion of PDB 1BC8 (ref. 60), PDB 1YO5 (ref. 61), PDB 1L3L (ref. 51) or PDB 2O4A (ref. 62) or (2) using the software X3DNA63, followed by a constrained Rosetta relax of the DNA structure.
Please note that 3DNA has been replaced by DSSR. The functionality for constructing B-DNA models, previously provided by 3DNA, is now directly available in DSSR via its fiber and rebuild modules.
In the preprint by Si et al. (2025), titled 'End-to-End Single-Stranded DNA Sequence Design with All-Atom Structure Reconstruction', DSSR is cited as follows:
Since ViennaRNA and NUPACK require secondary structures as input, we used DSSR35 to extract secondary structures from the corresponding ssDNA three-dimensional structures.
The above use cases are merely a sample of how DSSR is utilized in the scientific literature. It is reasonable to state that DSSR has emerged as a de facto standard tool within the field of nucleic acid structural bioinformatics. Overall, DSSR is a mature, robust, and efficient software product that is actively developed and maintained. I am committed to making DSSR synonymous with quality and value. Its unmatched functionality, usability, and support save users significant time and effort compared to alternative solutions.
DSSR is available free of charge for academic users. Additionally, it has been integrated into other high-profile bioinformatics resources, including NAKB, PDB-redo, and N•ESPript.
References
- Geng A, Roy R, Ganser L, Li L, Al-Hashimi HM. Revealing hidden protonated conformational states in RNA dynamic ensembles. Nucleic Acids Research. 2025;53:gkaf1366. https://doi.org/10.1093/nar/gkaf1366.
- Gordan R, Wasserman H, Chi B, Bohm K, Duan M, Sahay H, et al. High-throughput characterization of transcription factors that modulate UV damage formation and repair at single-nucleotide resolution. 2025. https://doi.org/10.21203/rs.3.rs-8197218/v1.
- Kubaney A, Favor A, McHugh L, Mitra R, Pecoraro R, Dauparas J, et al. RNA sequence design and protein–DNA specificity prediction with NA-MPNN. 2025. https://doi.org/10.1101/2025.10.03.679414.
- Glasscock CJ, Pecoraro RJ, McHugh R, Doyle LA, Chen W, Boivin O, et al. Computational design of sequence-specific DNA-binding proteins. Nat Struct Mol Biol. 2025;32:2252–61. https://doi.org/10.1038/s41594-025-01669-4.
- Si Y, Xu Y, Chen L. End-to-end single-stranded DNA sequence design with all-atom structure reconstruction. 2025. https://doi.org/10.64898/2025.12.05.692525.
While reading DNAproDB: an expanded database and web-based tool for structural analysis of DNA–protein complexes, I noticed SNAP and DSSR being mentioned. The detailed citations are as below:
Information about individual nucleotide–residue interactions is also provided, such as hydrogen bonding, interaction geometry (based on SNAP (10)), buried solvent accessible surface areas and identification of the interacting residue and nucleotide moieties …
DNAproDB assigns a geometry for every nucleotide–residue interaction identified using SNAP, a component of the 3DNA program suite (10). The residues for which probabilities are shown are those with planar side chains so that a stacking conformation can be defined.
Base pairing and base stacking between nucleotides are identified using the program DSSR (20).
SNAP and DSSR are two (relatively) new programs in the 3DNA software suite. As the author, I am always glad to see them being cited explicitly in literature. The fact that SNAP and DSSR are cited together by DNAproDB, however, is especially significant. I am aware of the initial version of DNAproDB, but I definitely like the updated one better. This is what I recently wrote in response to a question on the 3DNA Forum:
Regarding DNA-protein interactions in general, you may want to have a look of DNAproDB from the Remo Rohs laboratory. A new paper has just been published in NAR, ‘DNAproDB: an expanded database and web-based tool for structural analysis of DNA–protein complexes’.
I’ve no doubt that SNAP and DSSR would be widely used in applications related to DNA/RNA structural bioinformatics. DSSR (to a lesser extent, SNAP) represents my view of what a scientific software tool should be.

Recently I read the article Topology-based classification of tetrads and quadruplex structures in Bioinformatics by Popenda et al. In this work, the authors proposed an ONZ classification scheme of G-tetrads in intramolecular G-quadruplexes (G4) as shown below (Fig. 2 in the publication):
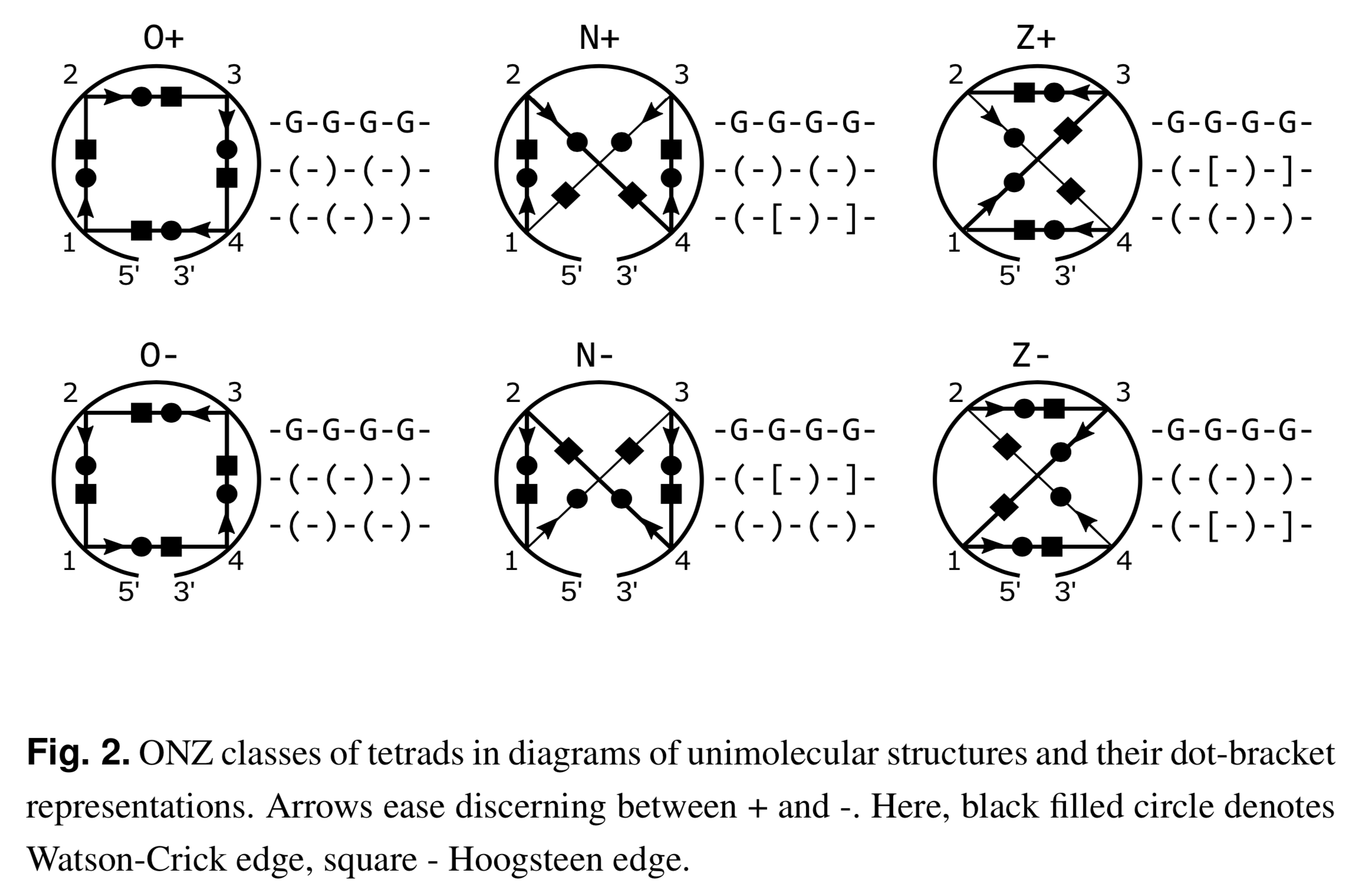
I am glad to find that DSSR has been used as a component in their computational tool ElTetrado to automatically identify and classify tetrads and quadruplexes.
Structures from both sets were analysed using self-implemented programs along with DSSR software from the 3DNA suite (Lu et al. (2015)). From DSSR, we acquired the information about base pairs and stacking.
I like the ONZ classification scheme: it is simple in concept yet provides a new perspective for the topologies of G-tetrads in intramolecular G4 structures. So I implemented the idea in DSSR v1.9.8-2019oct16, with this feature available via the --g4-onz option. Note that ElTetrado, according to the authors, is applicable to ONZ classifications of general types of tetrads and quadruplexes. The DSSR implementation of ONZ classifications, on the other hand, is strictly limited to G-tetrads in intramolecular G4 structures.
The DSSR ONZ classification results match the ones reported in Figs. 1, 5, and 6 of the Popenda et al. paper. For example, for PDB entry 6H1K (Fig. 6), the relevant results with the --g4-onz option and without it are listed below:
# x3dna-dssr -i=6h1k.pdb --g4-onz
List of 3 G-tetrads
1 glyco-bond=s--- groove=w--n planarity=0.149 type=planar Z- nts=4 GGGG A.DG1,A.DG20,A.DG16,A.DG27
2 glyco-bond=-sss groove=w--n planarity=0.136 type=planar Z+ nts=4 GGGG A.DG2,A.DG19,A.DG15,A.DG26
3 glyco-bond=--s- groove=-wn- planarity=0.307 type=other O+ nts=4 GGGG A.DG17,A.DG21,A.DG25,A.DG28
# ---------------------------------------
# x3dna-dssr -i=6h1k.pdb
# without option --g4-onz
List of 3 G-tetrads
1 glyco-bond=s--- groove=w--n planarity=0.149 type=planar nts=4 GGGG A.DG1,A.DG20,A.DG16,A.DG27
2 glyco-bond=-sss groove=w--n planarity=0.136 type=planar nts=4 GGGG A.DG2,A.DG19,A.DG15,A.DG26
3 glyco-bond=--s- groove=-wn- planarity=0.307 type=other nts=4 GGGG A.DG17,A.DG21,A.DG25,A.DG28
With the --json option, the ONZ classification results are always available. An example is shown below for PDB entry 6H1K (Fig. 6):
# x3dna-dssr -i=6h1k.pdb --json | jq -c '.G4tetrads[] | [.nts_long, .topo_class]'
["A.DG1,A.DG20,A.DG16,A.DG27","Z-"]
["A.DG2,A.DG19,A.DG15,A.DG26","Z+"]
["A.DG17,A.DG21,A.DG25,A.DG28","O+"]

I recently read a short communication by Pavel Afonine, titled phenix.hbond: a new tool for annotation hydrogen bonds in the July 2019 issue of the Computational Crystallography Newsletter (CCN). It appears that every bioinformatics tool (e.g., PyMOL or Jmol) has its own implementation of an algorithm on calculating H-bonds, one of the fundamental stabilizing forces of proteins and DNA/RNA structures. So does 3DNA/DSSR, as noted in my 2014-04-11 blogpost Get hydrogen bonds with DSSR.
Both DSSR and SNAP have the --get-hbond option, and they use the same underlying algorithm. However, the default output from the two programs differs: DSSR reports the H-bonds within nucleic acids, and SNAP covers only those at the DNA/RNA-protein interface. Using the PDB entry 1oct as an example, Running DSSR on it with the --get-hbond option gives 33 H-bonds in the DNA duplex, while SNAP reports 38 H-bonds at the DNA-protein interface. By design, the default output caters for the most-common use case of each program.
Under the scene, however, there exist variations in the seemingly simple --get-hbond option. One can attach text ‘nucleic’ (or ‘nuc’, ‘nt’), as in --get-hbond-nucleic, to output H-bonds within nucleic acids. Similarly, --get-hbond-protein (or ‘amino’, ‘aa’) would output H-bonds within proteins. Not surprisingly, the --get-hbond-nt-aa option would list H-bonds in nucleic acids and proteins, including those at their interface. These variations apply to both DSSR and SNAP, even though some are redundant with the default.
Notably, in combination with --json, the --get-hbond option by default would output all H-bonds, as if --get-hbond-nt-aa has been set. For PDB entry 1oct, DSSR or SNAP would report 208 H-bonds. Moreover, the JSON output has a residue_pair field for each identified H-bond, with values like "nt:nt", "nt:aa", or "aa:aa". Using 1oct as an example,
# x3dna-dssr -i=1oct.pdb --get-hbond --json | jq '.hbonds[0]'
{
"index": 1,
"atom1_serNum": 34,
"atom2_serNum": 608,
"donAcc_type": "standard",
"distance": 3.304,
"atom1_id": "O6@A.DG202",
"atom2_id": "N4@B.DC230",
"atom_pair": "O:N",
"residue_pair": "nt:nt"
}
# x3dna-dssr -i=1oct.pdb --get-hbond --json | jq '.hbonds[60]'
{
"index": 61,
"atom1_serNum": 462,
"atom2_serNum": 1187,
"donAcc_type": "standard",
"distance": 3.692,
"atom1_id": "O2@B.DT223",
"atom2_id": "NH2@C.ARG102",
"atom_pair": "O:N",
"residue_pair": "nt:aa"
}
# x3dna-dssr -i=1oct.pdb --get-hbond --json | jq '.hbonds[100]'
{
"index": 101,
"atom1_serNum": 791,
"atom2_serNum": 818,
"donAcc_type": "standard",
"distance": 2.871,
"atom1_id": "N@C.THR26",
"atom2_id": "OD2@C.ASP29",
"atom_pair": "N:O",
"residue_pair": "aa:aa"
}
In the above three cases, using SNAP instead of DSSR would give the same results.
Also, one can take advantage of the residue_pair value to filter H-bonds by type. For example, the following command would extract only H-bonds at the DNA-protein interface (38 occurrences, same as the number noted above):
x3dna-snap -i=1oct.pdb --get-hbond --json | jq '.hbonds[] | select(.residue_pair=="nt:aa")'
Back to the phenix.hbond tool, the author noted that:
Running phenix.hbond requires atomic model in PDB or mmCIF format with all hydrogen atoms added, as well as ligand restraint files if the model contains unknown to the library items.
While there is no particular reason why this should not work for all bio-macromolecules, currently phenix.hbond is only optimized and tested to work with proteins, which is the limitation that will be removed in future.
In contrast, the H-bond identification algorithm in DSSR/SNAP does not require hydrogen atoms. In fact, hydrogen atoms are simply ignored if they exist. As shown above, the H-bond method as implemented in DSSR/SNAP works for DNA, RNA, protein, or their complexes. This does not necessarily mean that the 3DNA way is superior to other similar tools. It just works well in my hand, and it may serve as a pragmatic choice for other users.

Recently I noticed two new citations to DSSR, an integrated software tool for dissecting the spatial structure of RNA. One is from the Yesselman et al. article Computational design of three-dimensional RNA structure and function in Nature Nanotechnology, and the other is from the Wang et al. article 3dRNA v2.0: An Updated Web Server for RNA 3D Structure Prediction in International Journal of Molecular Sciences.
Yesselman et al. has used DSSR in RNAMake for building the motif library. The relevant section is as follows:
We processed each RNA structure to extract every motif with Dissecting the Spatial Structure of RNA (DSSR)54 with the following command:
x3dna-dssr –i file.pdb –o file_dssr.out
We manually checked each extracted motif to confirm that it was the correct type, as DSSR sometimes classifies tertiary contacts as higher-order junctions and vice versa. For each motif collected from DSSR, we ran the X3DNA find_pair and analyze programs to determine the reference frame for the first and last base pair of each motif to allow for the alignment between motifs:
find_pair file.pdb 2> /dev/null stdout | analyze stdin >& /dev/null
It is worth noting the sentence that “DSSR sometimes classifies tertiary contacts as higher-order junctions and vice versa.” Presumably. the authors are referring to the inclusion of ‘isolated canonical pairs’ in junctions by default in DSSR. Overall, the default DSSR settings follow the most common practice in RNA literature. In the meantime, I am aware that the community may not agree on every detail. Thus DSSR provide many options (documented or otherwise) to cater for other potential use cases. See the Stems of junction structure have only one base pair and Junction definition threads on the 3DNA Forum for two examples. In the long run, DSSR is likely to help consolidate RNA nomenclature that can be applied in a pragmatic, consistent manner.
Note also that DSSR provides the reference frame of each identified base pair via the JSON option. Using 1ehz as an example, the following command provides detailed information about base pairs:
x3dna-dssr -i=1ehz.pdb --json --more | jq .pairs
In the 3dRNA 2.0 paper, DSSR is cited as below. This is the first time DSSR is integrated in the 3dRNA pipeline.
The predicted structures are built from the sequence and secondary structure, while the former is obtained from their native structures fetched from PDB (https://www.rcsb.org/), and the latter is calculated from DSSR (Dissecting the Spatial Structure of RNA) [39].

In the PDB, the ligand identifiers 5MC and 5CM all refer to 5-methylcytosine, but differ in the sugar moieties the base is attached to. Chemically, 5CM is 5-methyl-2’-deoxycytidine-5’-monophosphate as in DNA, and 5MC is 5-methylcytidine-5’-monophosphate. See the molecular images shown below.
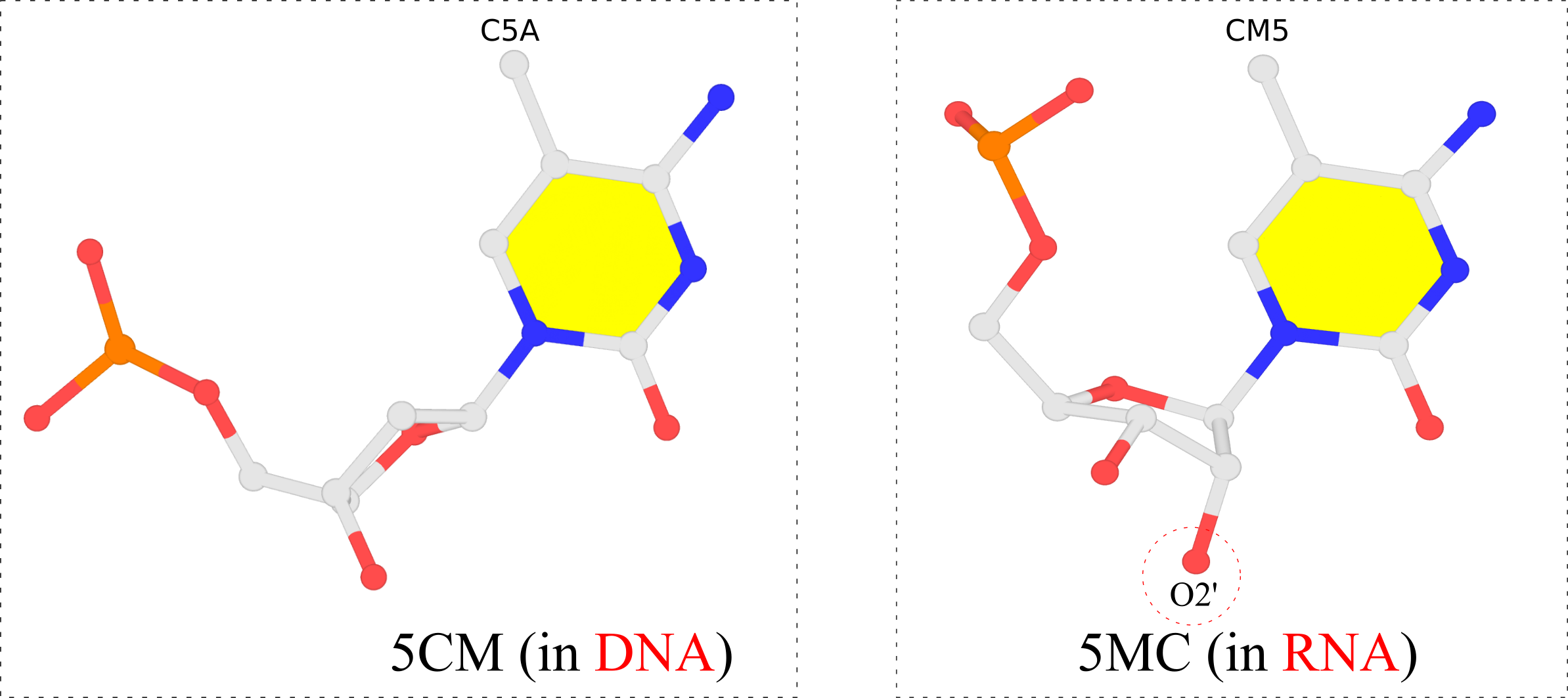
The 5-methyl group is named C5A in 5CM and CM5 in 5MC, respectively, for non-obvious reasons other than conventions. For comparison, the methyl-group in thymine of DNA is named C7, as for example in PDB id 355d. It is worth noting that DSSR is able to handle all such variations in atom or residue names.

I recently came across a Bioinformatics article VeriNA3d: an R package for nucleic acids data mining by Gallego et al. from IRB Barcelona. VeriNA3d can perform dataset analysis, single-structure analysis, and exploratory data analyses, with an emphasis on complex RNA structures. I am glad to see the DSSR is one of the third-party utilities that have been integrated into VeriNA3d, as shown below
VeriNA3d offers integration with third-party utilities such as the non-redundant lists of RNA structures (Leontis and Zirbel, 2012), the eRMSD suggested to compare RNA structures (Bottaro et al., 2014), a wrapper to the DSSR (Dissecting the Spatial Structure of RNA) software (Lu et al., 2015) and query functions to access the PDBe REST API (Velankar et al., 2016).
I browsed the GitLab repository and read through the supplemental documents. Clearly, VeriNA3d is a handy tool for the R community to perform RNA 3D structural analyses.
To DSSR users, Section “9 The dssr wrapper: getting the base pairs” of the supplemental PDF “VeriNA3d: introduction and use cases” is particularly relevant. The three paragraphs (with minor edits) are excerpted below:
The DSSR software (Dissecting the Spatial Structure of RNA) (Lu, Bussemaker, and Olson 2015) represents an invaluable resource to handle RNA structures. Some of the functions of veriNA3d overlap with the functionalities of DSSR, and both applications provide unique different features. We implement a wrapper to execute DSSR directly from R and get the best of both worlds in one place.
Note that installing veriNA3d does not automatically install DSSR, since we don’t redistribute third-party software. Before any user can use our wrapper, the dssr function, DSSR should be installed separately. To address this installation we redirect you to the DSSR manual, where anyone can find the specific instructions for their system. Once DSSR is installed and working in your computer, you will also be able to use it with our wrapper. If the DSSR executable (named x3dna-dssr) is in your path, dssr will find it automatically. If the wrapper does not find it, you can still use it specifying the absolute path to the executable with the argument exefile. Find more information running ?dssr.
One of the DSSR capabilities that users might be interested in is the detection and classification of base pairs. The following code shows a simple example. The output of the dssr wrapper is an object got from the json DSSR output. From R, json objects are parsed in the form of a tree of lists, with different types of information. Most of the interesting data is under the list models, sublist parameters, as shown herein.
I echo the authors’ policy of not redistributing third-party software with VeriNA3d. DSSR is under active development. Users should always visit the 3DNA Forum for downloading the latest version of DSSR, reporting bugs, and asking questions.
The R interface to DSSR (via JSON output) in VeriNA3d represents one of the intended use cases of DSSR’s many possible applications. No doubt DSSR is being increasingly integrated into other resources of RNA structural bioinformatics. Hopefully, more advanced DSSR features (than the detection and classification of base pairs) will also be widely appreciated in the future. Users would love DSSR better when they gain more experience in structural bioinformatics.

It is a great pleasure to see that our article Web 3DNA 2.0 for the analysis, visualization, and modeling of 3D nucleic acid structures has been highlighted in the cover page of the web server issue of NAR’19. According to the editor, This year, 331 proposals were submitted and 122, or 37%, were approved for manuscript submission. Of those approved, 94, or 77%, were ultimately accepted for publication. Overall, that corresponds to a ~28% acceptance rate.
The cover image and its caption are shown below. Moreover, details on how the cover image was created are available on the 3DNA Forum.

Caption: Examples of customized molecular models that can be generated with 3DNA: (top) a chromatin-like, nucleosome-decorated DNA with the structures of known histone-DNA assemblies placed at user-defined binding sites; (lower left) molecular schematic of a DNA trinucleotide diphosphate illustrating the base planes and reference frames used to construct and analyze 3D nucleic acid-containing structures; (lower right) customized single-stranded tRNA model built from a user-defined base sequence and a set of rigid-body parameters describing the desired placement of successive bases. Color code of base blocks: A, red; C, yellow; G, green; T, blue; U, cyan.

A paper titled DNA Conformational Changes Play a Force-Generating Role during Bacteriophage Genome Packaging has just been officially published in the Biophysical Journal (Volume 116, Issue 11, P2172-2180, June 04, 2019). I am glad to have the opportunity to collaborate with Kim Sharp, Gino Cingolani and Stephen Harvey on this interesting project that has big implications in understanding the mechanism of bacteriophage genome packaging. The abstract of the paper is shown below:
Motors that move DNA, or that move along DNA, play essential roles in DNA replication, transcription, recombination, and chromosome segregation. The mechanisms by which these DNA translocases operate remain largely unknown. Some double-stranded DNA (dsDNA) viruses use an ATP-dependent motor to drive DNA into preformed capsids. These include several human pathogens as well as dsDNA bacteriophages—viruses that infect bacteria. We previously proposed that DNA is not a passive substrate of bacteriophage packaging motors but is instead an active component of the machinery. We carried out computational studies on dsDNA in the channels of viral portal proteins, and they reveal DNA conformational changes consistent with that hypothesis. dsDNA becomes longer (“stretched”) in regions of high negative electrostatic potential and shorter (“scrunched”) in regions of high positive potential. These results suggest a mechanism that electrostatically couples the energy released by ATP hydrolysis to DNA translocation: The chemical cycle of ATP binding, hydrolysis, and product release drives a cycle of protein conformational changes. This produces changes in the electrostatic potential in the channel through the portal, and these drive cyclic changes in the length of dsDNA as the phosphate groups respond to the protein’s electrostatic potential. The DNA motions are captured by a coordinated protein-DNA grip-and-release cycle to produce DNA translocation. In short, the ATPase, portal, and dsDNA work synergistically to promote genome packaging.
Significantly, our work is highlighted in a “New and Notable” article, May the Road Rise to Meet You: DNA Deformation May Drive DNA Translocation by Paul Jardine (Volume 116, Issue 11, Pages 2060-2061, 4 June 2019):
Regardless of what drives conformational change in the portal, the idea that the linear DNA substrate is deformed in a way that makes it an energetic participant in its own movement opens new possibilities for how motors work. Large paddling or rotational motions by motor components may not be required if linear motion can be achieved by stretching or compressing the linear substrate, with rectified, cyclic conformational changes in the DNA rather than lever motions doing the work. If borne out by experiments, further simulation, and more structural information, this proposed mechanism may require a reappraisal of how we think about translocating motors.
For this project, I developed the x3dna-search program to survey similar fragments of single-stranded or double helical structures in the PDB.

After many years of efforts, it is a great pleasure to see our paper Effects of Noncanonical Base Pairing on RNA Folding: Structural Context and Spatial Arrangements of G·A Pairs published in ACS Biochemistry. The abstract is shown below:
Noncanonical base pairs play important roles in assembling the three-dimensional structures critical to the diverse functions of RNA. These associations contribute to the looped segments that intersperse the canonical double-helical elements within folded, globular RNA molecules. They stitch together various structural elements, serve as recognition elements for other molecules, and act as sites of intrinsic stiffness or deformability. This work takes advantage of new software (DSSR) designed to streamline the analysis and annotation of RNA three-dimensional structures. The multiscale structural information gathered for individual molecules, combined with the growing number of unique, well-resolved RNA structures, makes it possible to examine the collective features deeply and to uncover previously unrecognized patterns of chain organization. Here we focus on a subset of noncanonical base pairs involving guanine and adenine and the links between their modes of association, secondary structural context, and contributions to tertiary folding. The rigorous descriptions of base-pair geometry that we employ facilitate characterization of recurrent geometric motifs and the structural settings in which these arrangements occur. Moreover, the numerical parameters hint at the natural motions of the interacting bases and the pathways likely to connect different spatial forms. We draw attention to higher-order multiplexes involving two or more G·A pairs and the roles these associations appear to play in bridging different secondary structural units. The collective data reveal pairing propensities in base organization, secondary structural context, and deformability and serve as a starting point for further multiscale investigations and/or simulations of RNA folding.
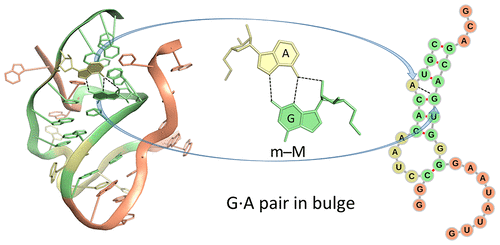
This work represents a multifaceted, fundamental application enabled by DSSR. Even at the base-pair (bp) level, DSSR provides unique features that complement the Leontis-Westhof (LW) notation of 12 geometric types.
At the review stage, we were asked by a referee to comment on the differences between DSSR and LW on bp classifications. The following paragraph in the “DISCUSSION” section of the paper is our response, expanded on the original writing that focused on DSSR’s capabilities:
Qualitative descriptions of noncanonical RNA base pairing, pioneered by Leontis and Westhof9,41 and linked in this work to the rigid-body parameters of interacting bases, have proven valuable in deciphering the connections between RNA primary, secondary, and tertiary structures. The present categorization is based on the positions of the hydrogen-bonded atoms with respect to a standard, embedded base reference frame30 defined in terms of an idealized Watson−Crick base pair. The major- and minor-groove base edges used here correspond in most cases to what are termed the Hoogsteen and sugar edges in the Leontis−Westhof scheme (one can compare the two classification schemes in Table S2). The + and − symbols introduced in 3DNA24 and DSSR27 unambiguously distinguish the relative orientations of the two bases. The trans and cis designations used in the earlier literature, however, are qualitative in nature and often uncertain. There are many “nc” (near cis, as in ncWW) and “nt” (near trans, as in ntSH) annotations listed in the RNA Structure Atlas; see, for example, the base-pair interactions in the sarcin−ricin domain of E. coli 23S rRNA found by entering PDB entry 1msy at http://rna.bgsu.edu/rna3dhub/pdb. The assignment of qualitative descriptors of RNA associations on the basis of atomic identity alone is generally not clear-cut. Numerical differences in the rigid-body parameters are critical to differentiating pairing schemes that share a common hydrogen bond, e.g., the G(N3)···A(N6) interaction found in m−WII and m−MI arrangements of G and A (Table 1 and Figures 4 and S3). The numerical data also provide a basis for following conformational transitions and may potentially be of value in making functional and other meaningful distinctions among RNA base pairs.
See also a recent thread Noncanonical base pair standards on the 3DNA Forum and the section titled “3.2.2 Base pairs” in the DSSR User Manual.


 March 2026
March 2026 February 2026
February 2026


