Recently, a 3DNA user asked on the Forum about how to perform mutations to 3-methyladenine. The user reported that the procedure described in the FAQ entry How can I mutate cytosine to 5-methylcytosine did not work for the case of 3-methyladenine. This ‘limitation’ is easily understandable: the 3DNA mutate_bases program must have knowledge of the target base, 3-methyladenine, to perform the mutation properly. The program works for the most common 5-methylcytosine mutations since the corresponding 5MC file (Atomic_5MC.pdb, in the standard base-reference frame) is already included within the 3DNA distribution. By supplying a similar file for the target base, mutate_bases runs the same for mutations to 5-methylcytosine (or other bases). This blog post outlines the procedure, using 3-methyladenine as an example.
A ligand name search for 5-methylcytosine on the RCSB PDB led to only two matched entries: 2X6F and 3MAG. The ligand id is 3MA. Since 3MAG has a better resolution (1.8 Å) than 2X6F (3.3 Å), its 3MA ligand was extracted from the corresponding PDB file (3MAG.pdb). The atomic coordinates, excluding those for the two hydrogens, are as below. Note that the 3-methyl carbon atom is named CN3.
HETATM 2960 N9 3MA A 600 16.587 14.258 22.170 1.00 49.87 N HETATM 2961 C4 3MA A 600 17.123 13.100 21.622 1.00 50.46 C HETATM 2962 N3 3MA A 600 16.877 11.811 22.009 1.00 50.37 N HETATM 2963 CN3 3MA A 600 15.983 11.363 23.063 1.00 50.41 C HETATM 2964 C2 3MA A 600 17.590 10.968 21.241 1.00 50.11 C HETATM 2965 N1 3MA A 600 18.422 11.217 20.224 1.00 49.27 N HETATM 2966 C6 3MA A 600 18.627 12.484 19.858 1.00 48.99 C HETATM 2967 N6 3MA A 600 19.426 12.709 18.829 1.00 46.12 N HETATM 2968 C5 3MA A 600 17.949 13.503 20.593 1.00 49.89 C HETATM 2969 N7 3MA A 600 17.929 14.900 20.488 1.00 49.84 N HETATM 2970 C8 3MA A 600 17.113 15.286 21.434 1.00 49.58 C
After running the 3DNA utility program std_base with options -fit -A, the corresponding atomic coordinates of 3MA are transformed to the standard base reference frame of adenine. The file must be named Atomic_3MA.pdb, and it has the following contents:
HETATM 1 N9 3MA A 1 -1.287 4.521 0.006 1.00 49.87 N HETATM 2 C4 3MA A 1 -1.262 3.133 0.004 1.00 50.46 C HETATM 3 N3 3MA A 1 -2.337 2.286 -0.009 1.00 50.37 N HETATM 4 CN3 3MA A 1 -3.743 2.648 -0.047 1.00 50.41 C HETATM 5 C2 3MA A 1 -1.905 1.013 0.001 1.00 50.11 C HETATM 6 N1 3MA A 1 -0.662 0.520 0.004 1.00 49.27 N HETATM 7 C6 3MA A 1 0.366 1.372 -0.003 1.00 48.99 C HETATM 8 N6 3MA A 1 1.588 0.867 -0.034 1.00 46.12 N HETATM 9 C5 3MA A 1 0.068 2.768 0.003 1.00 49.89 C HETATM 10 N7 3MA A 1 0.875 3.914 -0.003 1.00 49.84 N HETATM 11 C8 3MA A 1 0.026 4.909 -0.003 1.00 49.58 C
Note that in file Atomic_3MA.pdb, (1) the z-coordinates of the base atoms are close to zeros, (2) the ordering of atoms is as in the original ligand of 3MA shown above.
With Atomic_3MA.pdb in place (in the current working directory, or the $X3DNA/config folder), one can perform 3-methyladenine mutations using mutate_bases. For illustration purpose, let’s generate a B-form DNA with base sequence GACATGATTGCC using the 3DNA fiber program:
fiber -seq=GACATGATTGCC fiber-BDNA.pdb
To mutate A7 to 3MA, one needs to run mutate_bases as following:
mutate_bases "chain=A s=7 m=3MA" fiber-BDNA.pdb fiber-BDNA-A7to3MA.pdb
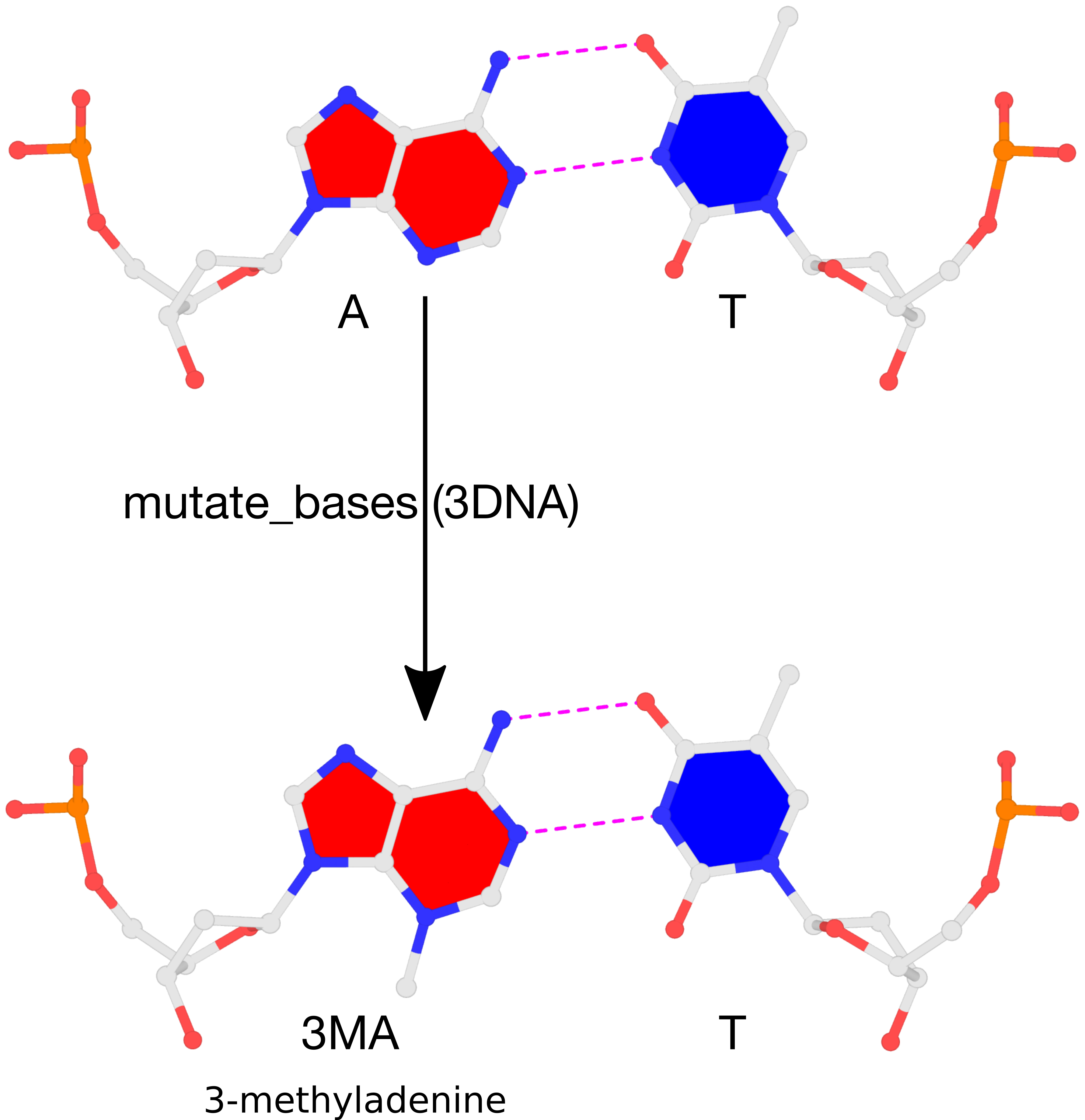
The result of the mutation is shown in the figure below. Note that the backbone has identical geometry as that before the mutation, and the mutated 3MA-T pair has exactly the same parameters (propeller/buckle etc) as the original A-T. These are the two defining features of the 3DNA mutate_bases program.
Please see the thread mutations to 3-methyladenine on the 3DNA Forum to download files fiber-BDNA.pdb and fiber-BDNA-A7to3MA.pdb.