Over the past couple of months, I’ve further enhanced the DSSR-derived structural features for Q-quadruplexes (G4). One was the implementation of the single descriptor of intramolecular canonical G4 structures with three connecting loops recently proposed by Dvorkin et al. The descriptor contains the number of guanines in the G4 stem, the type and relative direction of loops linking G-tracts of the stem, and the groove-widths associated with lateral loops. For example, PDB entry 2GKU (see the DSSR-enabled PyMOL schematic image below, Fig. 1A) has the following DSSR output.
List of 1 G4-stem
Note: a G4-stem is defined as a G4-helix with backbone connectivity.
Bulges are also allowed along each of the four strands.
stem#1[#1] layers=3 INTRA-molecular loops=3 descriptor=3(-P-Lw-Ln) note=hybrid-1(3+1) UUDU anti-parallel
1 glyco-bond=ss-s groove=-wn- mm(<>,outward) area=14.24 rise=3.58 twist=16.8 nts=4 GGGG A.DG3,A.DG9,A.DG17,A.DG21
2 glyco-bond=--s- groove=-wn- pm(>>,forward) area=13.12 rise=3.71 twist=25.9 nts=4 GGGG A.DG4,A.DG10,A.DG16,A.DG22
3 glyco-bond=--s- groove=-wn- nts=4 GGGG A.DG5,A.DG11,A.DG15,A.DG23
strand#1 U DNA glyco-bond=s-- nts=3 GGG A.DG3,A.DG4,A.DG5
strand#2 U DNA glyco-bond=s-- nts=3 GGG A.DG9,A.DG10,A.DG11
strand#3 D DNA glyco-bond=-ss nts=3 GGG A.DG17,A.DG16,A.DG15
strand#4 U DNA glyco-bond=s-- nts=3 GGG A.DG21,A.DG22,A.DG23
loop#1 type=propeller strands=[#1,#2] nts=3 TTA A.DT6,A.DT7,A.DA8
loop#2 type=lateral strands=[#2,#3] nts=3 TTA A.DT12,A.DT13,A.DA14
loop#3 type=lateral strands=[#3,#4] nts=3 TTA A.DT18,A.DT19,A.DA20
The descriptor=3(-P-Lw-Ln) means that the G4 structure has three layers of G-tetrads, connected via three loops: the first is the Propeller loop in anti-clockwise (negative) direction, then the Lateral loop passing a wide groove anti-clockwise, and finally another Lateral loop passing a narrow groove, also anti-clockwise. The DSSR symbols follow those of Dvorkin et al. but with capital letters L, P, and D for lateral, propeller, and diagonal loops instead of lower case letters (l, p, d) to avoid using subscript for groove-width info. So the 2GKU descriptor 3(-P-Lw-Ln) from DSSR corresponds to 3(-p-lw-ln) of Dvorkin et al.
The DSSR-enabled, PyMOL-rendered, block image in Fig. 1A makes the three G-tetrad layers (squared green blocks) immediately obvious. Other base identities and stacking interactions also become clear — for example, the A24 (in red) stacks on the top G-tetrad, and T1-A20 pair stacks with the bottom G-tetrad.
Two other PDB entries (2LOD and 2KOW) are illustrated in Fig. 1B and Fig. 1C. They have different topologies than 2GKU (Fig. 1A). DSSR is able to characterize all of them consistently.
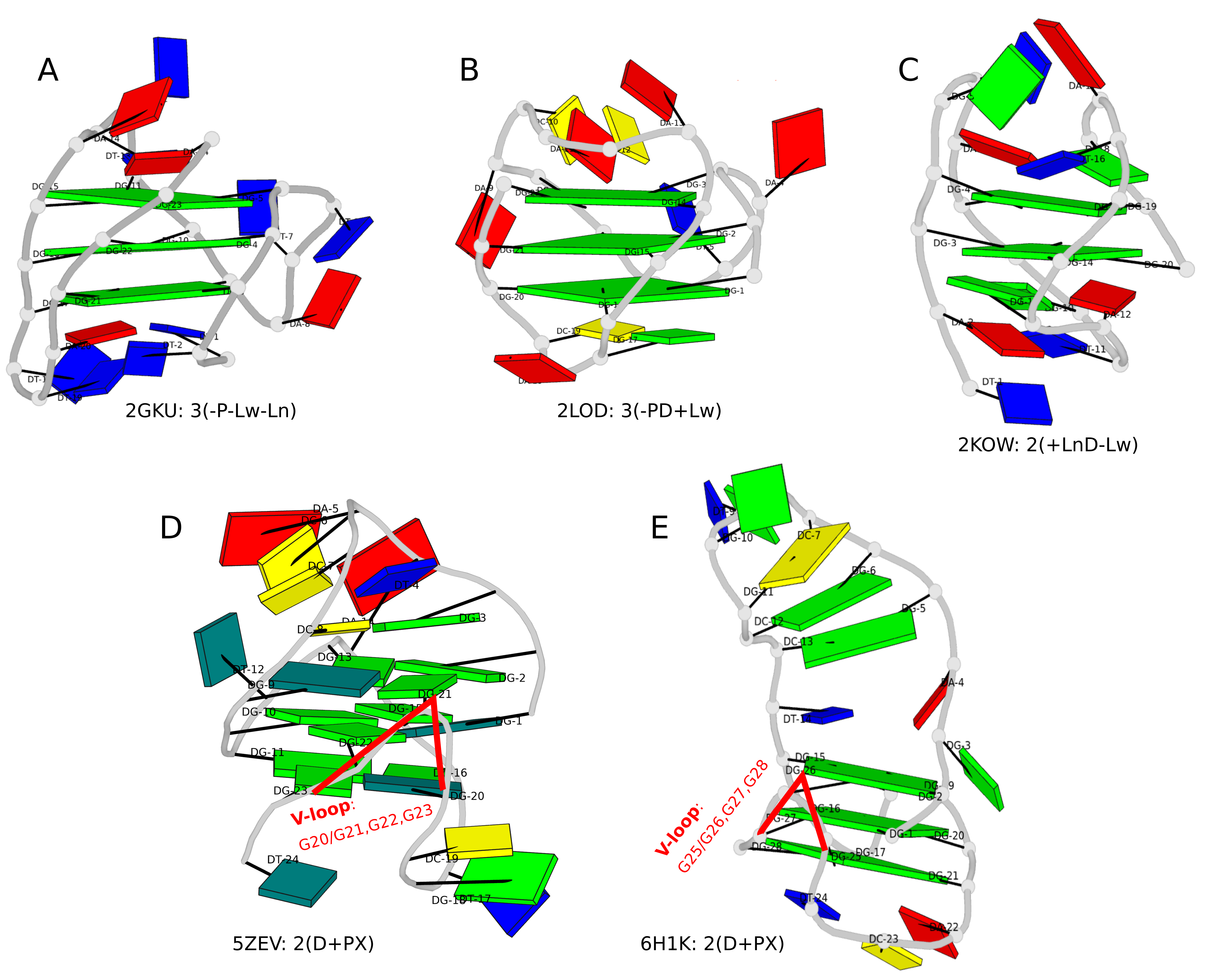
Figure 1. DSSR-enabled, PyMOL-rendered, block images of five G-quadruplexes. A in red, C in yellow, G (and G-tetrad) in green, and T in blue.
Another G4-related new feature in DSSR is the detection of V-shaped loops in noncanonical G4 structures where one of the four G-G columns (strands) that link adjacent G-tetrads is broken. Two of recent PDB examples with V-loops are shown in Fig. 1D (5ZEV) and Fig. 1E (6H1K). An excerpt of DSSR output for the PDB entry 6H1K is shown below.
List of 1 G4-helix
Note: a G4-helix is defined by stacking interactions of G4-tetrads, regardless
of backbone connectivity, and may contain more than one G4-stem.
helix#1[1] stems=[#1] layers=3 INTRA-molecular
1 glyco-bond=-sss groove=w--n mm(<>,outward) area=12.76 rise=3.47 twist=18.2 nts=4 GGGG A.DG2,A.DG19,A.DG15,A.DG26
2 glyco-bond=s--- groove=w--n pm(>>,forward) area=12.84 rise=3.07 twist=33.4 nts=4 GGGG A.DG1,A.DG20,A.DG16,A.DG27
3 glyco-bond=s--- groove=w--n nts=4 GGGG A.DG25,A.DG21,A.DG17,A.DG28
strand#1 DNA glyco-bond=-ss nts=3 GGG A.DG2,A.DG1,A.DG25
strand#2 DNA glyco-bond=s-- nts=3 GGG A.DG19,A.DG20,A.DG21
strand#3 DNA glyco-bond=s-- nts=3 GGG A.DG15,A.DG16,A.DG17
strand#4 DNA glyco-bond=s-- nts=3 GGG A.DG26,A.DG27,A.DG28
****************************************************************************
List of 1 G4-stem
Note: a G4-stem is defined as a G4-helix with backbone connectivity.
Bulges are also allowed along each of the four strands.
stem#1[#1] layers=2 INTRA-molecular loops=3 descriptor=2(D+PX) note=UD3(1+3) UDDD anti-parallel
1 glyco-bond=s--- groove=w--n mm(<>,outward) area=12.76 rise=3.47 twist=18.2 nts=4 GGGG A.DG1,A.DG20,A.DG16,A.DG27
2 glyco-bond=-sss groove=w--n nts=4 GGGG A.DG2,A.DG19,A.DG15,A.DG26
strand#1 U DNA glyco-bond=s- nts=2 GG A.DG1,A.DG2
strand#2 D DNA glyco-bond=-s nts=2 GG A.DG20,A.DG19
strand#3 D DNA glyco-bond=-s nts=2 GG A.DG16,A.DG15
strand#4 D DNA glyco-bond=-s nts=2 GG A.DG27,A.DG26
loop#1 type=diagonal strands=[#1,#3] nts=12 GAGGCGTGGCCT A.DG3,A.DA4,A.DG5,A.DG6,A.DC7,A.DG8,A.DT9,A.DG10,A.DG11,A.DC12,A.DC13,A.DT14
loop#2 type=propeller strands=[#3,#2] nts=2 GC A.DG17,A.DC18
loop#3 type=diag-prop strands=[#2,#4] nts=5 GACTG A.DG21,A.DA22,A.DC23,A.DT24,A.DG25
****************************************************************************
List of 2 non-stem G4 loops (INCLUDING the two terminal nts)
1 type=lateral helix=#1 nts=5 GACTG A.DG21,A.DA22,A.DC23,A.DT24,A.DG25
2 type=V-shaped helix=#1 nts=4 GGGG A.DG25,A.DG26,A.DG27,A.DG28
Note that here a new loop type (diag-prop) and topology description symbol (X) are introduced. In developing DSSR in general, and G4-related features in particular, I’ve always tried to follow conventions widely used by the community. Whereas inconsistency exists, I pick up the ones that are in line with other parts of DSSR. For unique DSSR features lacking outside references, I came up my own nomenclature. When DSSR becomes more widely used, it may serve to standardize G4 nomenclatures.
