It is a great pleasure to announce the publication of Web 3DNA 2.0 for the analysis, visualization, and modeling of 3D nucleic acid structures in Nucleic Acids Research (NAR). The paper will appear in the web server issue of NAR in July 2019. At nine-page in length and with several new structural parameters, this w3DNA 2.0 paper is certainly not a typical NAR web-server publication. It represents a significant contribution to the field of 3D nucleic acids structural bioinformatics, and will undoubtedly push the popularity of 3DNA to a new level.
The abstract is shown below:
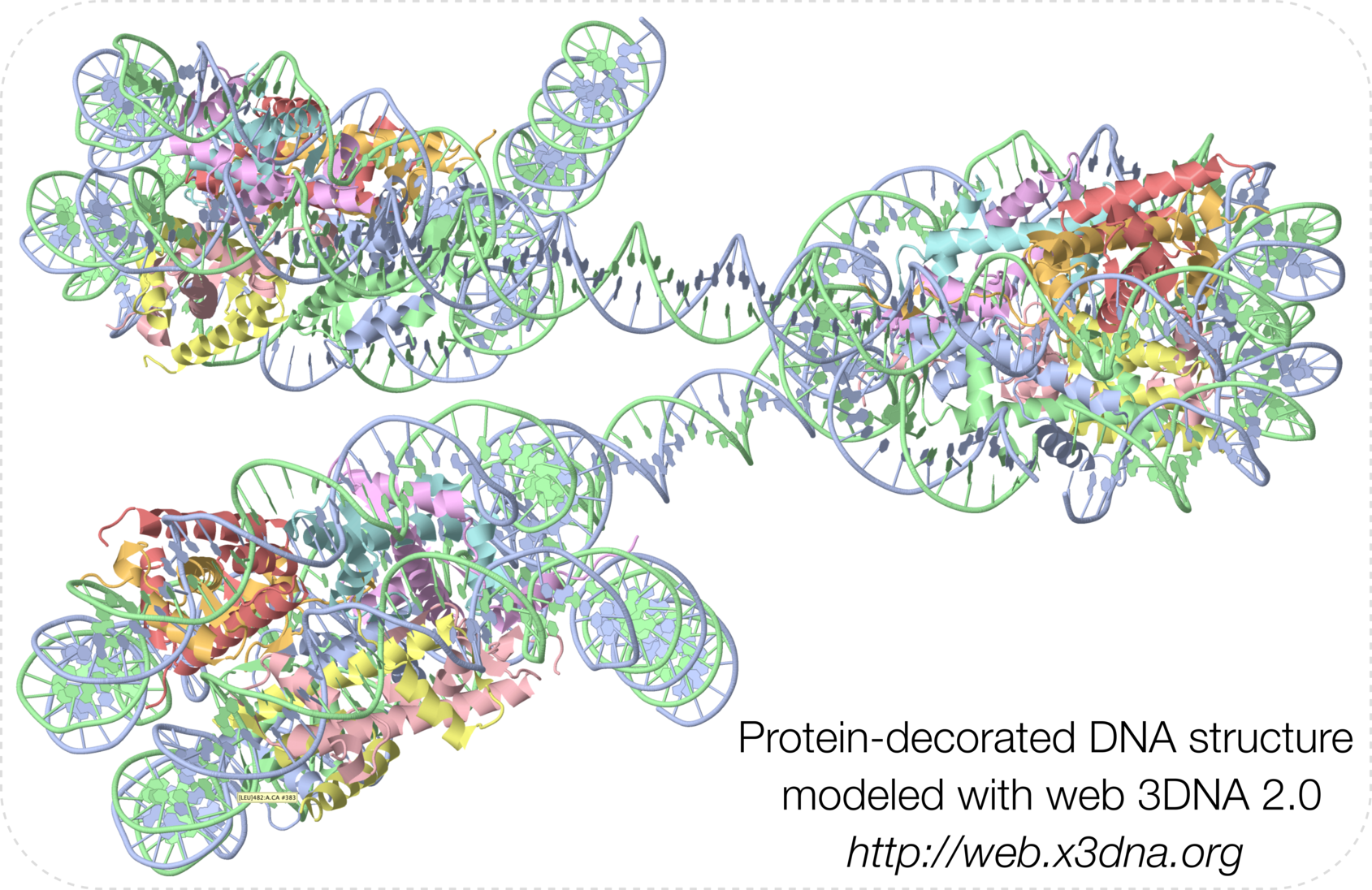
Web 3DNA (w3DNA) 2.0 is a significantly enhanced version of the widely used w3DNA server for the analysis, visualization, and modeling of 3D nucleic-acid-containing structures. Since its initial release in 2009, the w3DNA server has continuously served the community by making commonly-used features of the 3DNA suite of command-line programs readily accessible. However, due to the lack of updates, w3DNA has clearly shown its age in terms of modern web technologies and it has long lagged behind further developments of 3DNA per se. The w3DNA 2.0 server presented here overcomes all known shortcomings of w3DNA while maintaining its battle-tested characteristics. Technically, w3DNA 2.0 implements a simple and intuitive interface (with sensible defaults) for increased usability, and it complies with HTML5 web standards for broad accessibility. Featurewise, w3DNA 2.0 employs the most recent version of 3DNA, enhanced with many new functionalities, including: the automatic handling of modified nucleotides; a set of ‘simple’ base-pair and step parameters for qualitative characterization of non-Watson–Crick double- helical structures; new structural parameters that integrate the rigid base plane and the backbone phosphate group, the two nucleic acid components most reliably determined with X-ray crystallography; in silico base mutations that preserve the backbone geometry; and a notably improved module for building models of single-stranded RNA, double- helical DNA, Pauling triplex, G-quadruplex, or DNA structures ‘decorated’ with proteins. The w3DNA 2.0 server is freely available, without registration, at http://web.x3dna.org.
Moreover, details on reproducing our reported results are available in a dedicated section ‘web 3DNA 2.0 (http://web.x3dna.org)’ on the 3DNA Forum.
- Figure 1. Summary of web 3DNA 2.0
- Figure 2. New structural parameters
- Figure 3. Fiber models and mutations
- Supplemental PDF
- Graphical abstract (taken from Fig. 1E):