Via PDB weekly update, I recently came across PDB entry 6neb, which is solved by NMR and described as an “MYC promoter G-quadruplex with 1:6:1 loop length”. I downloaded the atomic coordinates of the entry and ran DSSR on it. Indeed, DSSR readily identifies a three-layered parallel G-quadruplex (G4) with three propeller-type loops of 1, 6 and 1 nucleotides (i.e., 1:6:1), as shown below.
List of 1 G4-stem
Note: a G4-stem is defined as a G4-helix with backbone connectivity.
Bulges are also allowed along each of the four strands.
stem#1[#1] layers=3 INTRA-molecular loops=3 descriptor=3(-P-P-P) note=parallel(4+0) UUUU parallel
1 glyco-bond=---- groove=---- WC-->Major nts=4 GGGG A.DG3,A.DG7,A.DG16,A.DG20
pm(>>,forward) area=14.54 rise=3.36 twist=24.7
2 glyco-bond=---- groove=---- WC-->Major nts=4 GGGG A.DG4,A.DG8,A.DG17,A.DG21
pm(>>,forward) area=9.67 rise=3.48 twist=30.3
3 glyco-bond=---- groove=---- WC-->Major nts=4 GGGG A.DG5,A.DG9,A.DG18,A.DG22
strand#1 U DNA glyco-bond=--- nts=3 GGG A.DG3,A.DG4,A.DG5
strand#2 U DNA glyco-bond=--- nts=3 GGG A.DG7,A.DG8,A.DG9
strand#3 U DNA glyco-bond=--- nts=3 GGG A.DG16,A.DG17,A.DG18
strand#4 U DNA glyco-bond=--- nts=3 GGG A.DG20,A.DG21,A.DG22
loop#1 type=propeller strands=[#1,#2] nts=1 A A.DA6
loop#2 type=propeller strands=[#2,#3] nts=6 TTTTAA A.DT10,A.DT11,A.DT12,A.DT13,A.DA14,A.DA15
loop#3 type=propeller strands=[#3,#4] nts=1 T A.DT19
I then read the associated paper titled Solution Structure of a MYC Promoter G-Quadruplex with 1:6:1 Loop Length lately published in the new, open-access ACS Omega journal. The reported structure 6neb has a 27-nt sequence (termed Myc1245) of bases 5’-TTGGGGAGGGTTTTAAGGGTGGGGAAT-3’. Myc1245 is based on the 27-nt long, purine-rich MycPu27 which has 5 tracts of guanines of G4-forming motif within the MYC promoter. In Myc1245, the third G-tract of MycPu27 has been replaced by TTTA, thus it uses only G-tracts 1, 2, 4, 5 for G4 formation. Previously, it was shown that Myc2345 (using G-tracts 2-5 of MycPu27) adopts a parallel G4 structure with three propeller loops of 1:2:1 nt length.
The MycPu27 sequence is representative of the G4-forming nuclease hypersensitive element (NHE III1) within the promoter region of the MYC oncogene. Formation of G4 structures suppresses MYC transcription, thus ligand-induced G4 stabilization in the DNA level is a promising strategy for cancer therapy. The NHE III1 motif can fold into multiple G4 structures depending on factors such as protein binding. The paper on 6neb illustrates that nucleolin, a protein shown to bind MYC G4 and repress MYC transcription, preferably binds the 1:6:1 loop length conformer than the 1:2:1 conformer (the major form under physiological conditions).
The DSSR analysis of 6neb shows that the two G-tetrad steps have different overlapping areas and twist angles. The top step comprising G3 and G4 (Fig. 1A) has better stacking interactions (14.5 Å2) and smaller twist (25º) than the bottom step containing G4 and G5 (9.7 Å2 and 30º, respectively).
area=14.54 rise=3.36 twist=24.7 area=9.67 rise=3.48 twist=30.3
The analysis characterizes the T1–A15 pair as a reverse Hoogsteen pair (rHoogsteen), which is distinct from the T+A Hoogsteen pair. In DSSR, the rHoogsteen pair is of type M–N (anti-parallel), whilst the Hoogsteen pair is of type M+N (parallel). With the local base-reference frames attached (Fig. 1B), it is easy to visualize that the z-axis of T1 is pointing out of the base-pair plane, and the z-axis of A15 is pointing inwards. See also my blog post Hoogsteen and reverse Hoogsteen base pairs.
Fig. 1C shows the ATG-triad automatically identified by DSSR. As is clear in Fig. 1A, the ATG-triad stacks on the 3-layered G4 structure on the 5’ side. Moreover, with color-coded base blocks (G in green, T in blue, and A in red), the two stacks (T10–T11, and T12–T13–A14) in the 6-nt central propeller loop is immediately obvious.
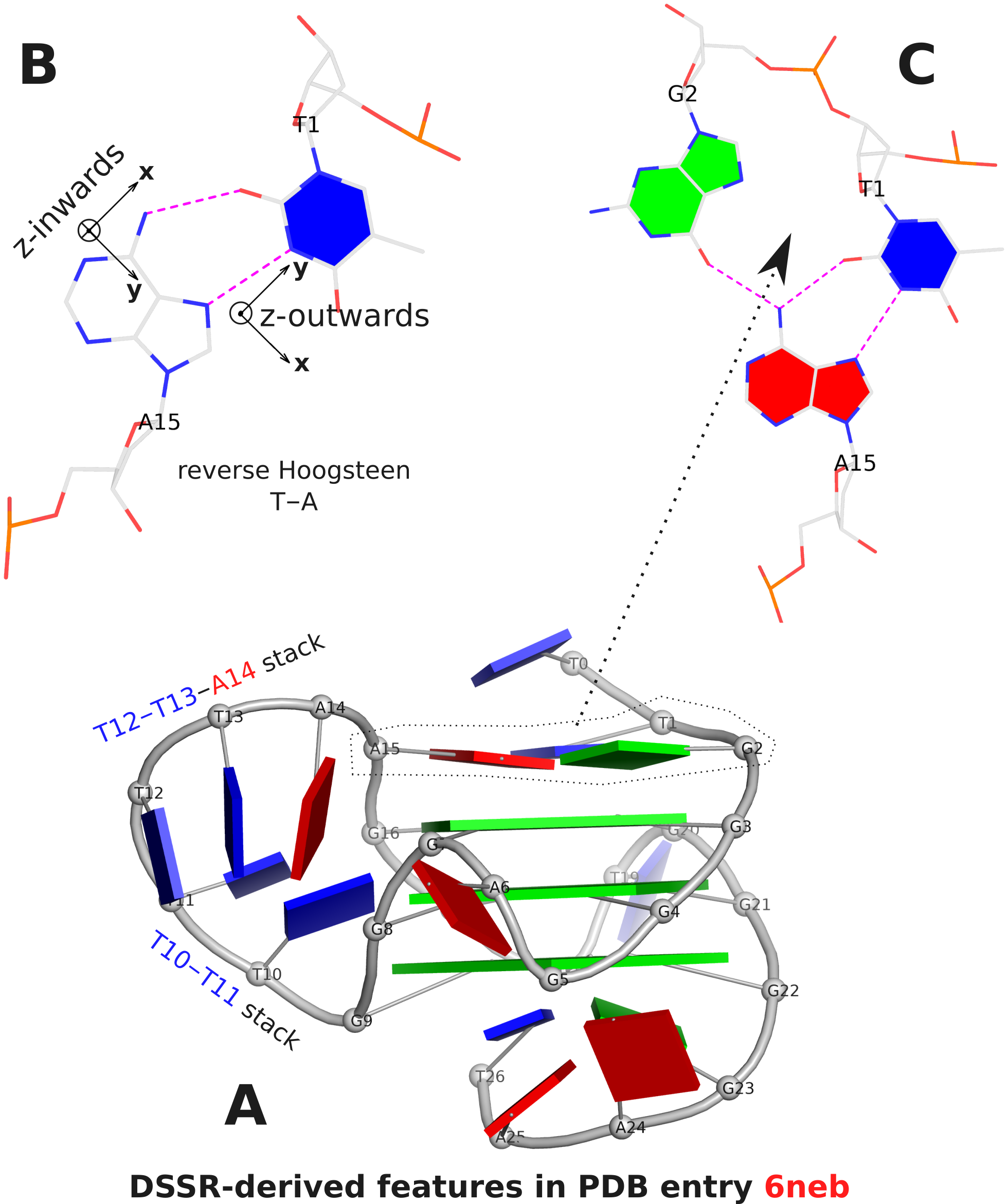
Figure 1. DSSR-derived structural features in PDB entry 6neb. The images were created using DSSR and PyMOL.
In the 6neb paper, the author stated that “The central loop of 6 nt connects the outer tetrads by spanning the G-core. A single nucleotide is the minimal length of this structural motif, so the five additional residues can significantly increase the loop’s conformational flexibility.” (p.2536) It is worth noting that in PDB entry 2m53, described in G-rich VEGF aptamer with locked and unlocked nucleic acid modifications exhibits a unique G-quadruplex fold, it was observed that:
An unprecedented all parallel-stranded monomeric G-quadruplex with three G-quartet planes exhibits several unique structural features. Five consecutive guanine residues are all involved in G-quartet formation and occupy positions in adjacent DNA strands, which are bridged with a no-residue propeller-type loop.
The G4 structure is polymorphic. It seems every imaginable or even unexpected form is possible, depending on the context.
