Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. [Nucleic Acids Res 48: e74(https://doi.org/10.1093/nar/gkaa426)).
See the 2020 paper titled "DSSR-enabled innovative schematics of 3D nucleic acid structures with PyMOL" in Nucleic Acids Research and the corresponding Supplemental PDF for details. Many thanks to Drs. Wilma Olson and Cathy Lawson for their help in the preparation of the illustrations.
Details on how to reproduce the cover images are available on the 3DNA Forum.

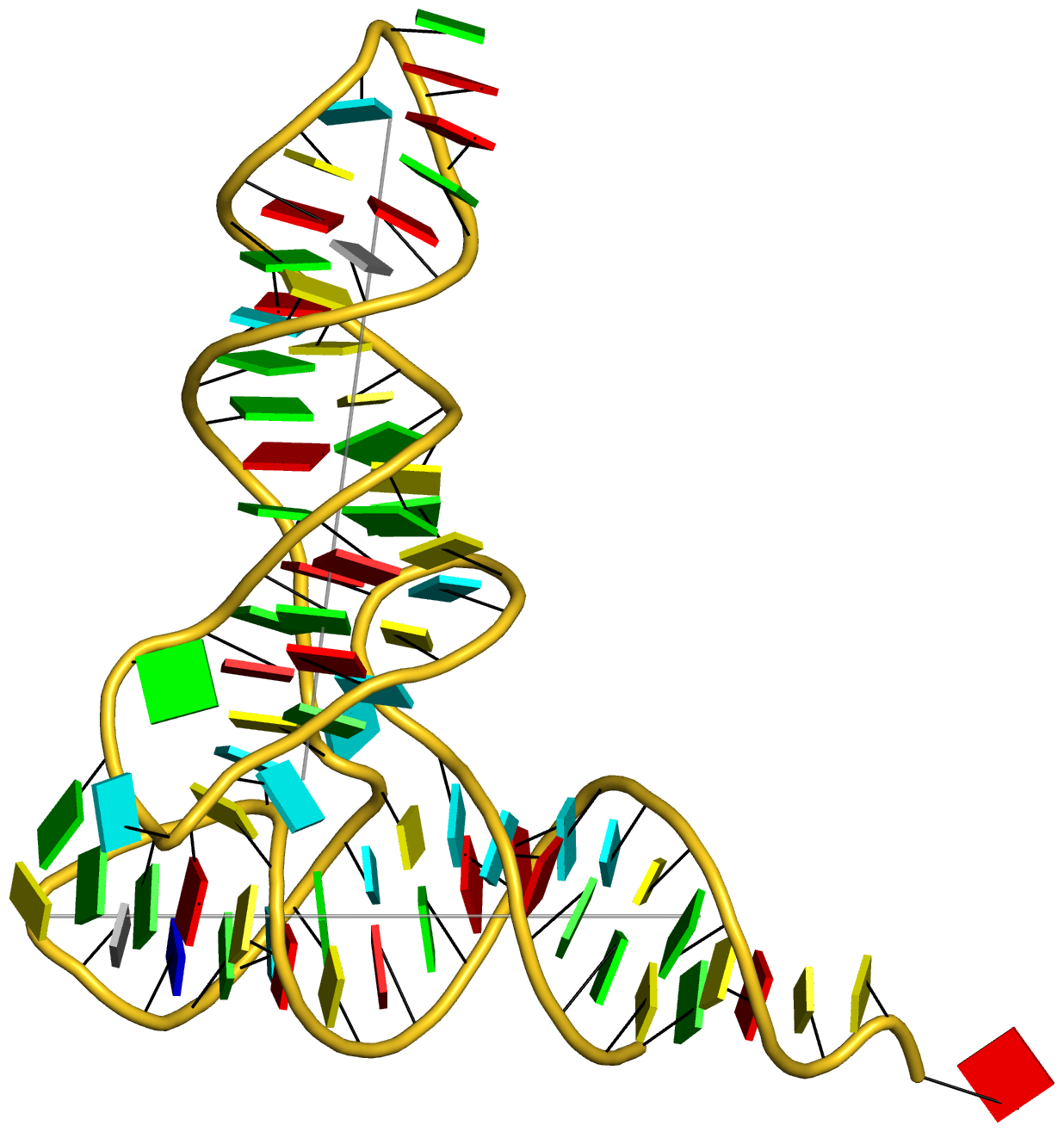
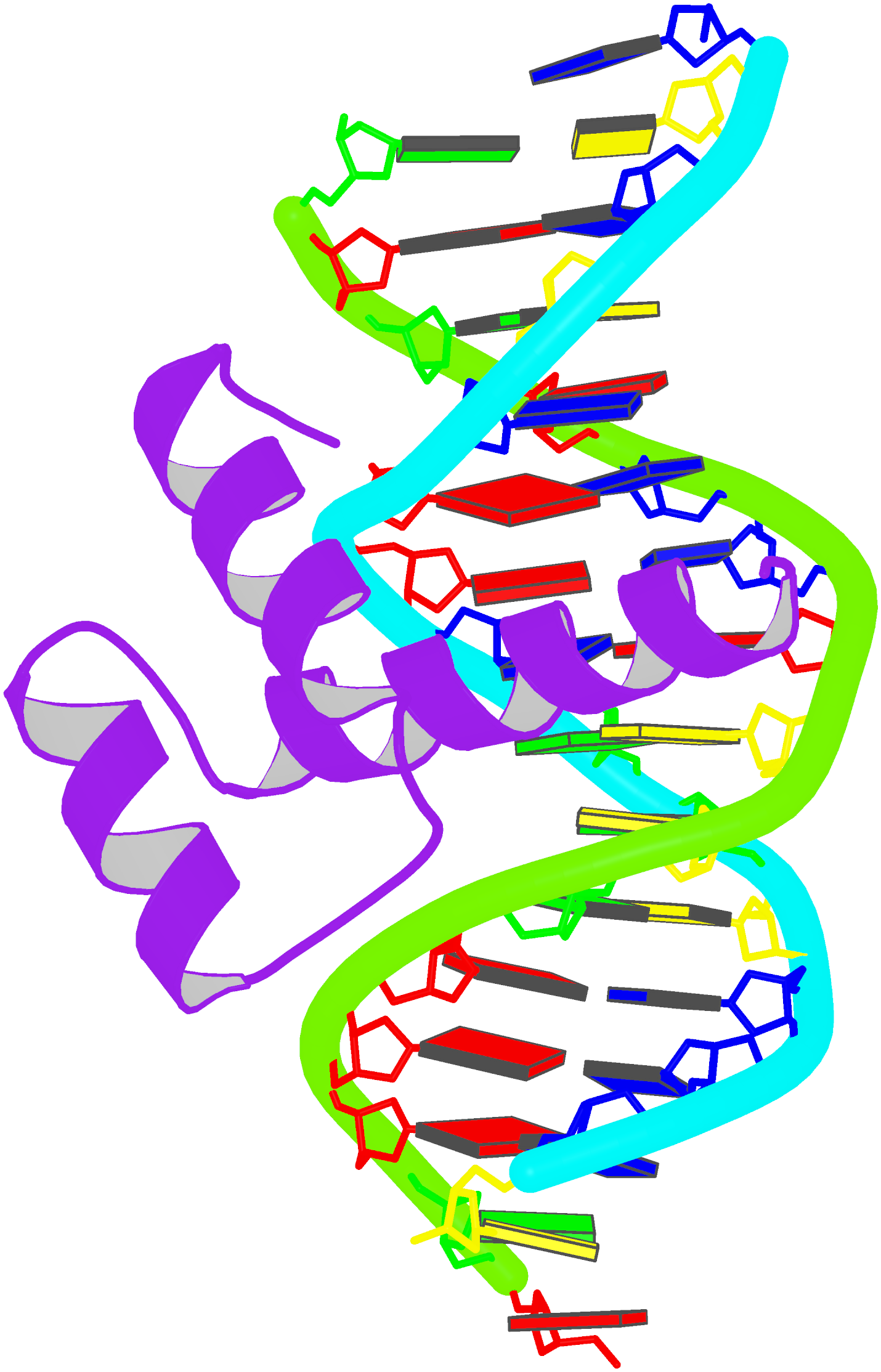
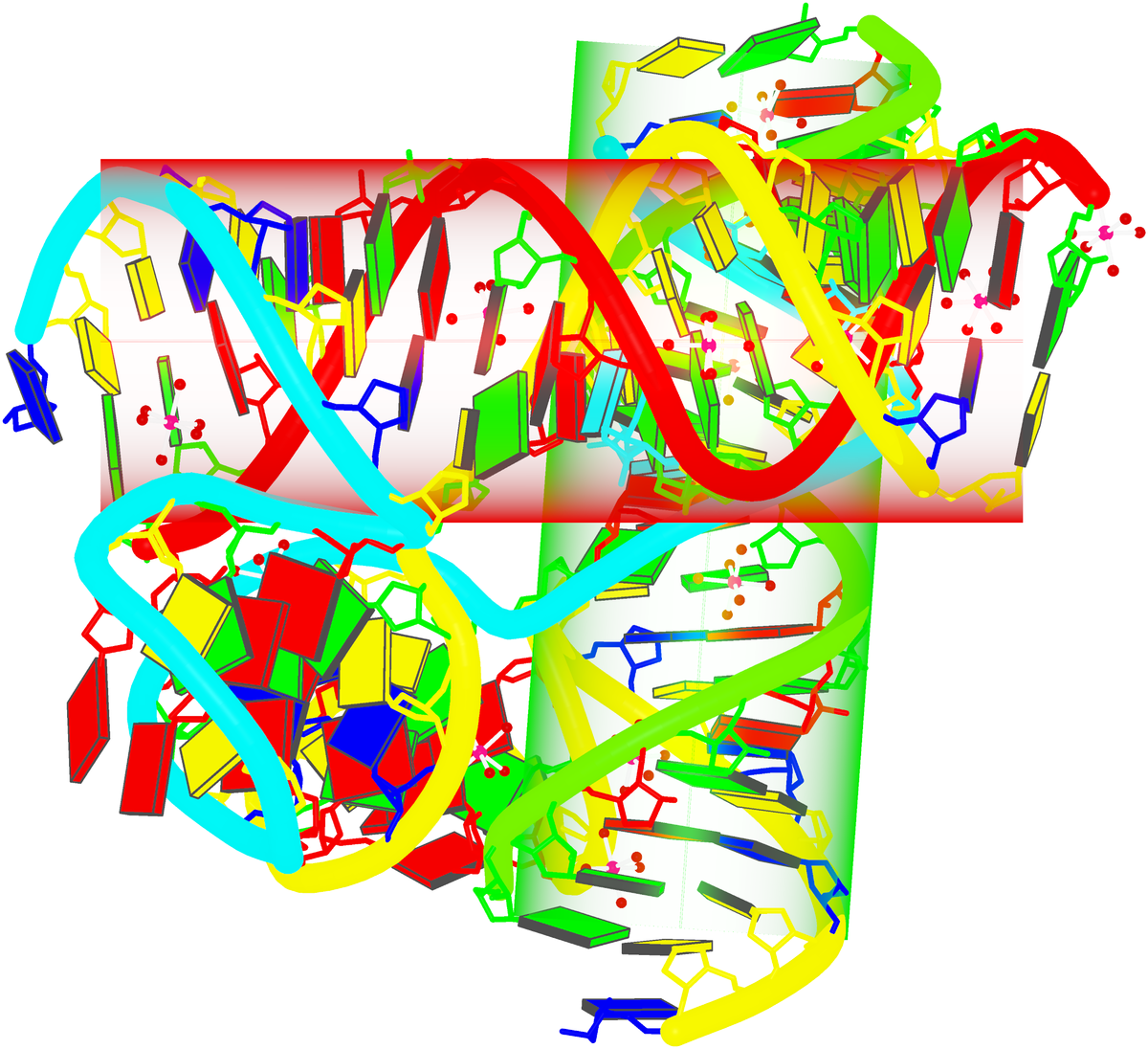
Structure of a group II intron ribonucleoprotein in the pre-ligation state (PDB id: 8T2R; Xu L, Liu T, Chung K, Pyle AM. 2023. Structural insights into intron catalysis and dynamics during splicing. Nature 624: 682–688). The pre-ligation complex of the Agathobacter rectalis group II intron reverse transcriptase/maturase with intron and 5′-exon RNAs makes it possible to construct a picture of the splicing active site. The intron is depicted by a green ribbon, with bases and Watson-Crick base pairs represented as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; the 5′-exon is shown by white spheres and the protein by a gold ribbon. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

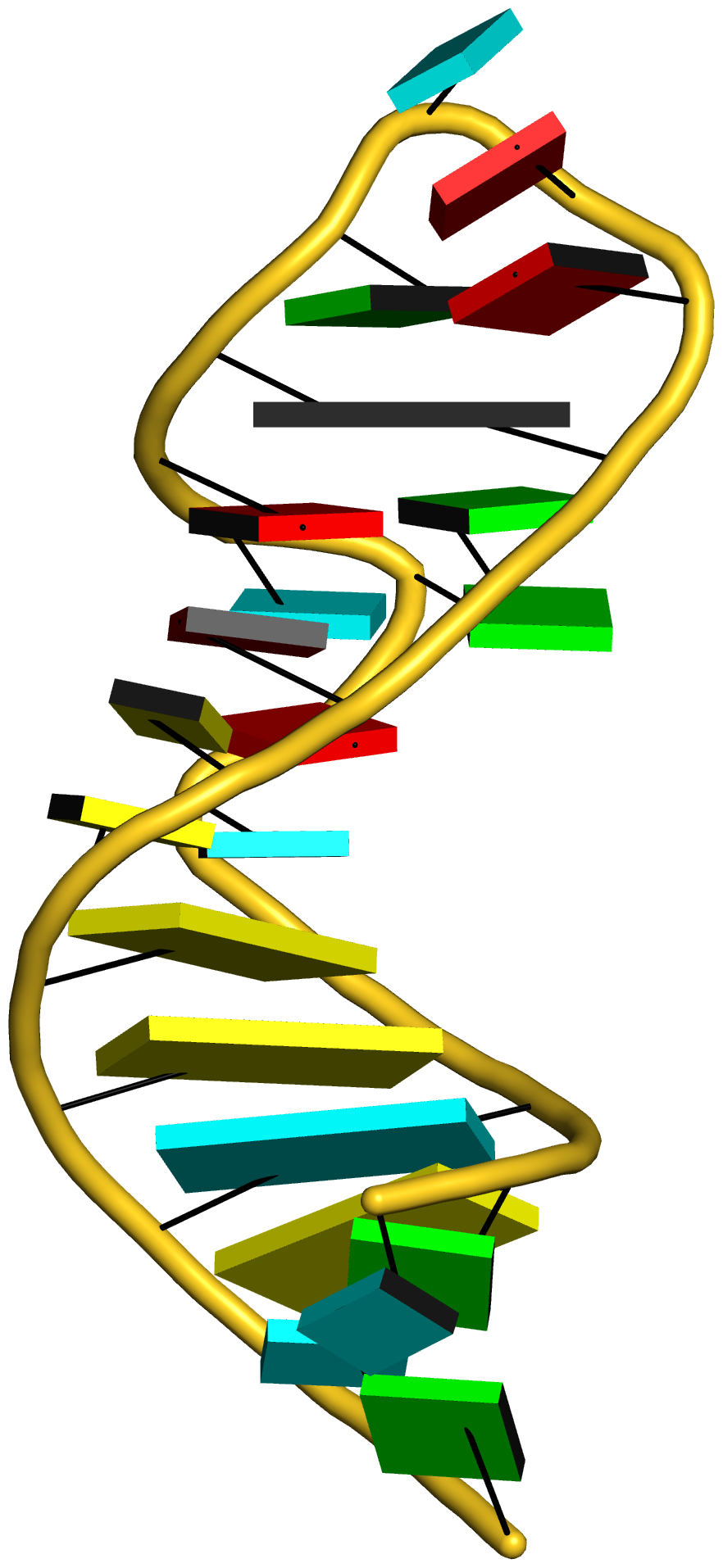
Complex of terminal uridylyltransferase 7 (TUT7) with pre-miRNA and Lin28A (PDB id: 8OPT; Yi G, Ye M, Carrique L, El-Sagheer A, Brown T, Norbury CJ, Zhang P, Gilbert RJ. 2024. Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs. Nat Struct Mol Biol 31: 1426–1438). The RNA-binding pluripotency factor LIN28A invades and melts the RNA and affects the mechanism of action of the TUT7 enzyme. The RNA backbone is depicted by a red ribbon, with bases and Watson-Crick base pairs represented as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; TUT7 is represented by a gold ribbon and LIN28A by a white ribbon. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

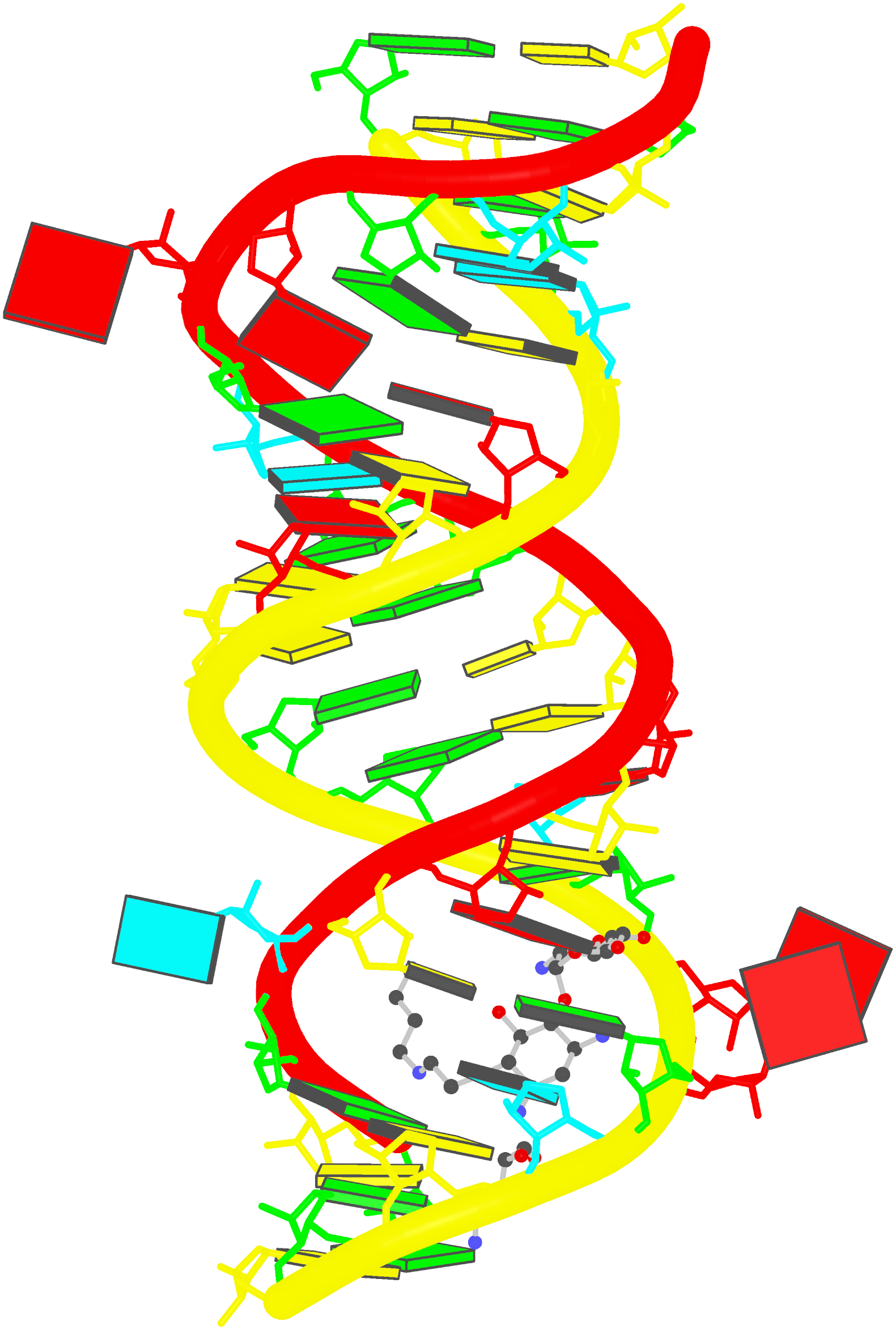
Cryo-EM structure of the pre-B complex (PDB id: 8QP8; Zhang Z, Kumar V, Dybkov O, Will CL, Zhong J, Ludwig SE, Urlaub H, Kastner B, Stark H, Lührmann R. 2024. Structural insights into the cross-exon to cross-intron spliceosome switch. Nature 630: 1012–1019). The pre-B complex is thought to be critical in the regulation of splicing reactions. Its structure suggests how the cross-exon and cross-intron spliceosome assembly pathways converge. The U4, U5, and U6 snRNA backbones are depicted respectively by blue, green, and red ribbons, with bases and Watson-Crick base pairs shown as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; the proteins are represented by gold ribbons. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

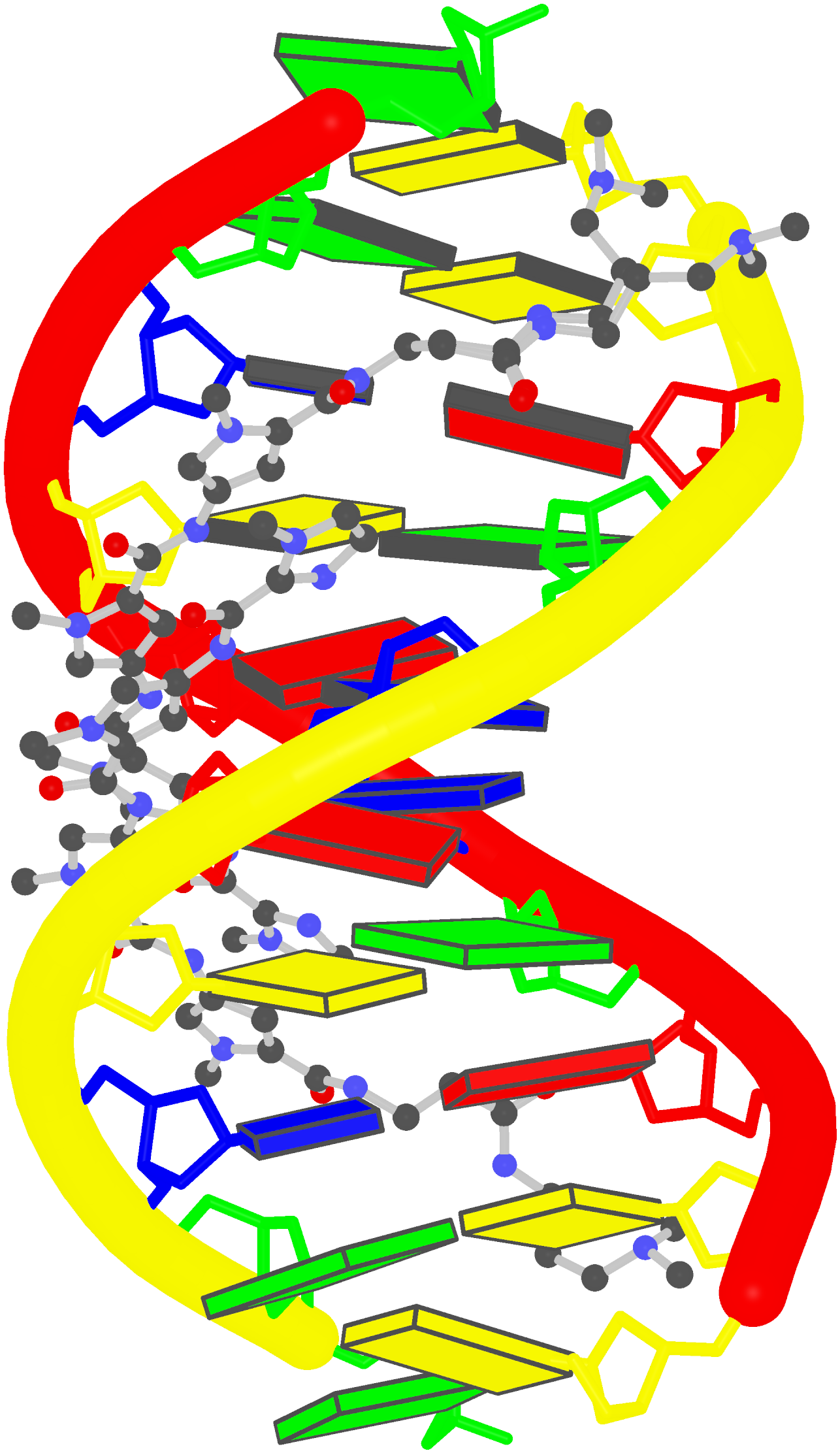
Structure of the Hendra henipavirus (HeV) nucleoprotein (N) protein-RNA double-ring assembly (PDB id: 8C4H; Passchier TC, White JB, Maskell DP, Byrne MJ, Ranson NA, Edwards TA, Barr JN. 2024. The cryoEM structure of the Hendra henipavirus nucleoprotein reveals insights into paramyxoviral nucleocapsid architectures. Sci Rep 14: 14099). The HeV N protein adopts a bi-lobed fold, where the N- and C-terminal globular domains are bisected by an RNA binding cleft. Neighboring N proteins assemble laterally and completely encapsidate the viral genomic and antigenomic RNAs. The two RNAs are depicted by green and red ribbons. The U bases of the poly(U) model are shown as cyan blocks. Proteins are represented as semitransparent gold ribbons. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Structure of the helicase and C-terminal domains of Dicer-related helicase-1 (DRH-1) bound to dsRNA (PDB id: 8T5S; Consalvo CD, Aderounmu AM, Donelick HM, Aruscavage PJ, Eckert DM, Shen PS, Bass BL. 2024. Caenorhabditis elegans Dicer acts with the RIG-I-like helicase DRH-1 and RDE-4 to cleave dsRNA. eLife 13: RP93979. Cryo-EM structures of Dicer-1 in complex with DRH-1, RNAi deficient-4 (RDE-4), and dsRNA provide mechanistic insights into how these three proteins cooperate in antiviral defense. The dsRNA backbone is depicted by green and red ribbons. The U-A pairs of the poly(A)·poly(U) model are shown as long rectangular cyan blocks, with minor-groove edges colored white. The ADP ligand is represented by a red block and the protein by a gold ribbon. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).
Moreover, the following 30 [12(2021) + 12(2022) + 6(2023)] cover images of the RNA Journal were generated by the NAKB (nakb.org).
Cover image provided by the Nucleic Acid Database (NDB)/Nucleic Acid Knowledgebase (NAKB; nakb.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).
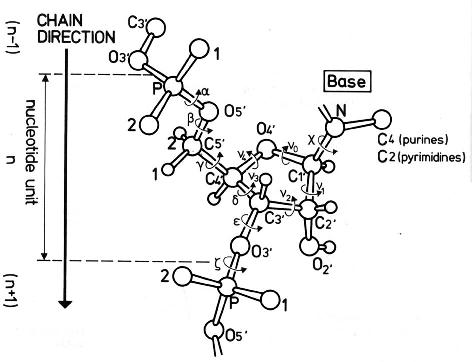
Backbone conformation of nucleic acid structures is most characterized by a set of 6 torsion angles (α, β, γ, δ, ε, and ζ) around the consecutive chemical bonds, chi (χ) quantifying the relative base/sugar orientation, plus the sugar pucker.
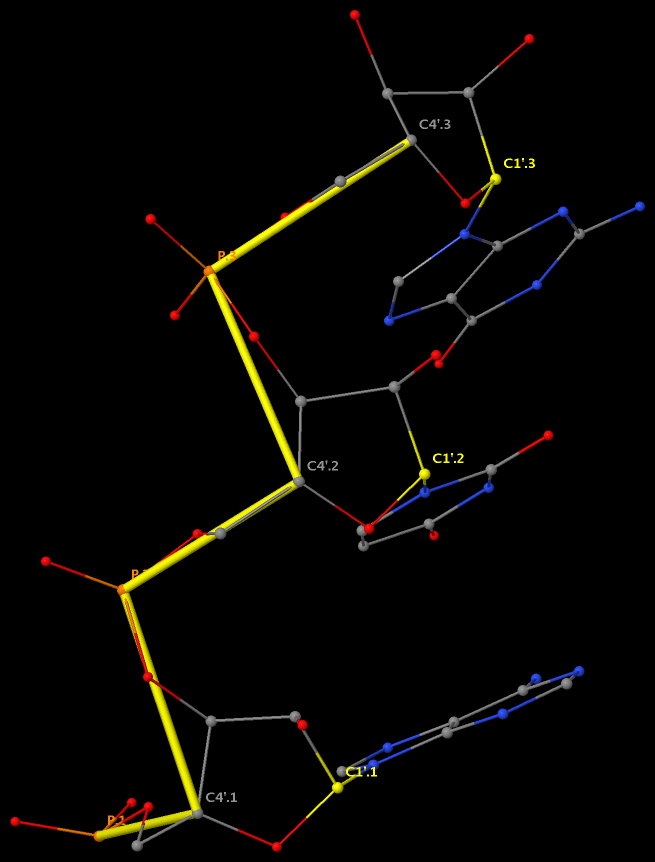
This large number of DNA/RNA backbone conformational parameters is in striking contrast to the two torsion angles (φ and ψ) in protein structures, routinely employed in Ramachandran plot. Over the years, the nucleic acid community has come up with simplified ways to represent DNA/RNA backbone conformation. Thus far, the most widely used one is the pseudo-torsion angles (See figure below) η: C4′(i-1)-P(i)-C4′(i)-P(i+1) and θ: P(i)-C4′(i)-P(i+1)-C4′(i+1).
The history of the P—C4′ virtual-bond concept and its application in RNA structure analysis have recently been reviewed by Pyle et al. in A new way to see RNA [Q Rev Biophys. 2011, 44(4), 433—466], where the following three contributions are highlighted:
- Olson (1980). Configurational statistics of polynucleotide chains. An updated virtual bond model to treat effects of base stacking., Macromolecules 13(3), 721—728.
- Malathi & Yathindra (1980). A novel virtual bond scheme to probe ordered and random coil conformations of nucleic acids: Configurational statistics of polynucleotide chains. Current Science, 49, 803—807.
- Duarte & Pyle (1998). Stepping through an RNA structure: A novel approach to conformational analysis. Journal of Molecular Biology, 284, 1465—1478.
More recently, Pyle et al. also employed a modified version of the pseudo-torsions, η′: C1′(i-1)-P(i)-C1′(i)-P(i+1) and θ′: P(i)-C1′(i)-P(i+1)-C1′(i+1), i.e., using C1′ instead of C4′, and found that:
The η′ and θ′ torsions are more suitable when interpreting crystallographic density because the C1′ atom is covalently bound to the nucleoside base and therefore can be more easily and accurately located within a low-resolution map.
While implementing the -torsion option to analyze to make it more explicit that 3DNA readily calculates conventional backbone torsion angles, I also take this opportunity to add the pseudo-torsion angles — η/θ and η′/θ′, among other new parameters. Moreover, while I am at it, I cannot help but also compute yet another set of pseudo-torsion angles: η″/θ″. Here, instead of C1′ or C4′, the origin of the base reference frame is employed; it can be taken as a _pseudo_-atom more accurately defined by the base plane than any real single atom.
The usefulness of η″/θ″, especially in comparison with η/θ and η′/θ′, remains to be determined. However, only η″/θ″ uniquely takes advantage of the two most accurately determined entities in a nucleic acid structure, the heavy phosphorus atom and the rigid base plane [see discussion (p.16) in the Richardson et al. MolProbity paper, Acta Cryst. (2010). D66, 12–21] Presumably, η″/θ″ provides a new perspective in RNA structural analysis by combining the backbone and the base.
Here is the pseudo-torsions for the yeast phenylalanine transfer RNA (6tna by simply running analyze -torsion=6tna.tor 6tna.pdb):
Pseudo (virtual) eta/theta torsion angles:
Note: eta: C4'(i-1)-P(i)-C4'(i)-P(i+1)
theta: P(i)-C4'(i)-P(i+1)-C4'(i+1)
eta': C1'(i-1)-P(i)-C1'(i)-P(i+1)
theta': P(i)-C1'(i)-P(i+1)-C1'(i+1)
eta": Borg(i-1)-P(i)-Borg(i)-P(i+1)
theta": P(i)-Borg(i)-P(i+1)-Borg(i+1)
base eta theta eta' theta' eta" theta"
1 A:...1_:[..G]G --- -126.6 --- -141.5 --- -130.4
2 A:...2_:[..C]C 167.8 -168.3 174.6 -152.5 -151.4 -115.4
3 A:...3_:[..G]G 160.4 -119.8 -171.9 -138.9 -123.6 -119.2
4 A:...4_:[..G]G 148.0 -164.2 162.1 -159.2 -154.4 -124.6
5 A:...5_:[..A]A 168.7 -137.6 -175.9 -137.8 -129.5 -115.0
6 A:...6_:[..U]U 171.8 -145.7 -172.5 -140.5 -131.3 -124.7
7 A:...7_:[..U]U -151.0 -47.8 -136.0 -58.6 -117.7 -30.2
8 A:...8_:[..U]U 160.9 159.7 -161.0 -163.6 -144.2 178.0
9 A:...9_:[..A]A -137.0 -48.6 -158.1 -108.9 161.5 -104.7
10 A:..10_:[2MG]g 33.1 -135.8 93.4 -134.6 134.1 -113.0
11 A:..11_:[..C]C 167.2 -138.3 -179.4 -137.7 -142.4 -118.7
12 A:..12_:[..U]U 165.5 -120.7 -179.3 -128.0 -145.8 -106.7
13 A:..13_:[..C]C 174.1 -173.6 -165.5 179.6 -120.9 -180.0
14 A:..14_:[..A]A 173.0 -144.0 172.7 -132.4 177.6 -72.7
15 A:..15_:[..G]G 154.7 110.6 -176.2 85.5 -97.7 -76.9
16 A:..16_:[H2U]u 76.3 94.1 65.3 119.7 -152.8 -123.8
17 A:..17_:[H2U]u -36.7 -79.6 -50.7 -136.6 -142.7 -159.0
18 A:..18_:[..G]G -9.7 -166.8 41.7 -158.6 28.9 -120.4
19 A:..19_:[..G]G -131.6 -35.8 -122.9 -67.8 -104.3 -10.5
20 A:..20_:[..G]G 160.9 -93.2 -161.6 -98.9 -174.1 -112.3
21 A:..21_:[..A]A -83.6 152.5 -72.8 155.7 -59.1 155.4
22 A:..22_:[..G]G 164.1 169.4 160.0 -178.5 159.1 -157.6
23 A:..23_:[..A]A 177.6 -148.5 -174.5 -142.7 -154.5 -114.3
24 A:..24_:[..G]G 167.2 -98.9 -171.7 -128.6 -127.6 -99.1
25 A:..25_:[..C]C 151.6 -153.5 167.3 -140.8 -137.7 -84.8
26 A:..26_:[M2G]g 156.2 -137.4 -175.2 -135.2 -100.0 -104.2
27 A:..27_:[..C]C 166.2 -145.5 -177.9 -140.4 -129.1 -116.8
28 A:..28_:[..C]C 164.7 -140.5 175.8 -145.3 -152.7 -123.4
29 A:..29_:[..A]A 161.2 -145.3 175.7 -144.9 -142.0 -126.0
30 A:..30_:[..G]G -173.5 -120.3 -158.4 -133.2 -126.6 -94.4
31 A:..31_:[..A]A 169.8 -153.1 177.7 -140.4 -124.5 -81.5
32 A:..32_:[OMC]c 154.4 -126.8 -178.7 -131.3 -104.1 -128.0
33 A:..33_:[..U]U 170.0 -103.9 -179.9 -152.7 -164.6 143.6
34 A:..34_:[OMG]g -4.7 -123.7 41.8 -124.8 31.6 -99.6
35 A:..35_:[..A]A 163.5 -104.3 176.9 -127.9 -137.5 -128.2
36 A:..36_:[..A]A 175.9 173.6 180.0 -167.7 -156.4 -118.3
37 A:..37_:[.YG]g 166.8 -131.7 -174.5 -133.0 -115.1 -82.9
38 A:..38_:[..A]A 167.7 -121.6 -175.7 -114.3 -109.9 -79.9
39 A:..39_:[PSU]P 168.3 -146.8 -160.2 -146.4 -98.6 -116.5
40 A:..40_:[5MC]c 160.6 -138.7 174.0 -141.8 -139.7 -126.5
41 A:..41_:[..U]U 164.8 -161.4 175.9 -152.3 -150.5 -117.6
42 A:..42_:[..G]G 174.3 -140.9 -170.3 -145.4 -129.1 -121.3
43 A:..43_:[..G]G 169.6 -159.0 -176.2 -154.9 -133.7 -133.1
44 A:..44_:[..A]A 174.0 -121.5 -174.2 -122.0 -143.1 -74.9
45 A:..45_:[..G]G 174.4 -132.5 -166.2 -128.1 -101.8 -128.9
46 A:..46_:[7MG]g -112.8 -113.4 -127.2 -138.3 -139.8 -152.1
47 A:..47_:[..U]U -63.2 -53.8 -1.1 -92.0 22.8 -124.7
48 A:..48_:[..C]C -84.7 59.6 -20.1 8.9 19.3 -104.5
49 A:..49_:[5MC]c -56.8 -140.1 -29.9 -143.6 98.1 -125.4
50 A:..50_:[..U]U 173.6 -146.4 -178.3 -140.6 -147.6 -117.8
51 A:..51_:[..G]G 160.8 -148.1 -178.6 -150.7 -140.7 -121.9
52 A:..52_:[..U]U 164.9 -144.0 175.8 -143.5 -139.9 -114.3
53 A:..53_:[..G]G 168.2 -140.9 -171.1 -144.0 -121.6 -117.3
54 A:..54_:[5MU]u 167.0 -131.1 178.3 -124.9 -139.9 -77.0
55 A:..55_:[PSU]P 167.6 -114.2 -172.8 -155.6 -113.0 146.0
56 A:..56_:[..C]C 35.0 -121.5 52.6 -126.2 26.5 -83.8
57 A:..57_:[..G]G 168.4 -148.1 -177.1 -131.1 -115.4 -111.7
58 A:..58_:[1MA]a -136.3 -133.3 -106.5 -176.7 -105.3 149.6
59 A:..59_:[..U]U 23.0 -130.9 33.0 -115.4 48.2 -68.2
60 A:..60_:[..C]C -163.6 -54.3 -123.2 -76.4 -79.6 -36.4
61 A:..61_:[..C]C 125.5 -153.3 169.7 -144.7 -153.8 -123.4
62 A:..62_:[..A]A 172.5 -139.3 -177.0 -137.6 -150.7 -114.6
63 A:..63_:[..C]C 165.8 -146.6 -178.5 -149.8 -139.2 -127.8
64 A:..64_:[..A]A 164.7 -144.9 176.5 -145.8 -145.3 -118.1
65 A:..65_:[..G]G 170.4 -152.3 -175.5 -151.5 -132.3 -122.1
66 A:..66_:[..A]A 168.0 -152.0 -177.4 -150.2 -133.0 -118.7
67 A:..67_:[..A]A 170.9 -141.8 -178.4 -140.4 -134.8 -123.1
68 A:..68_:[..U]U 164.8 -135.1 -178.9 -137.9 -143.7 -95.2
69 A:..69_:[..U]U 168.2 -154.9 -174.3 -157.1 -112.2 -144.8
70 A:..70_:[..C]C 160.6 -153.2 170.7 -153.5 -164.4 -125.1
71 A:..71_:[..G]G 161.8 -144.3 172.1 -143.1 -145.7 -124.2
72 A:..72_:[..C]C 176.7 -136.4 -169.3 -134.5 -134.9 -87.1
73 A:..73_:[..A]A 160.6 -142.8 -179.7 -139.7 -112.8 -104.4
74 A:..74_:[..C]C -176.9 -115.9 -163.1 -115.4 -117.2 -68.7
75 A:..75_:[..C]C 169.8 80.9 -170.0 74.9 -108.5 -91.3
76 A:..76_:[..A]A --- --- --- --- --- ---

In nucleic acid structures, the chi (χ) torsion angle is about the glycosidic bond (N-C1′) that connects the sugar and the A/C/G/T/U bases (or their modified variants). Specifically, for pyrimidines (C, T and U), χ is defined by O4′-C1′-N1-C2; and for purines (A and G) by O4′-C1′-N9-C4 (see figure below).

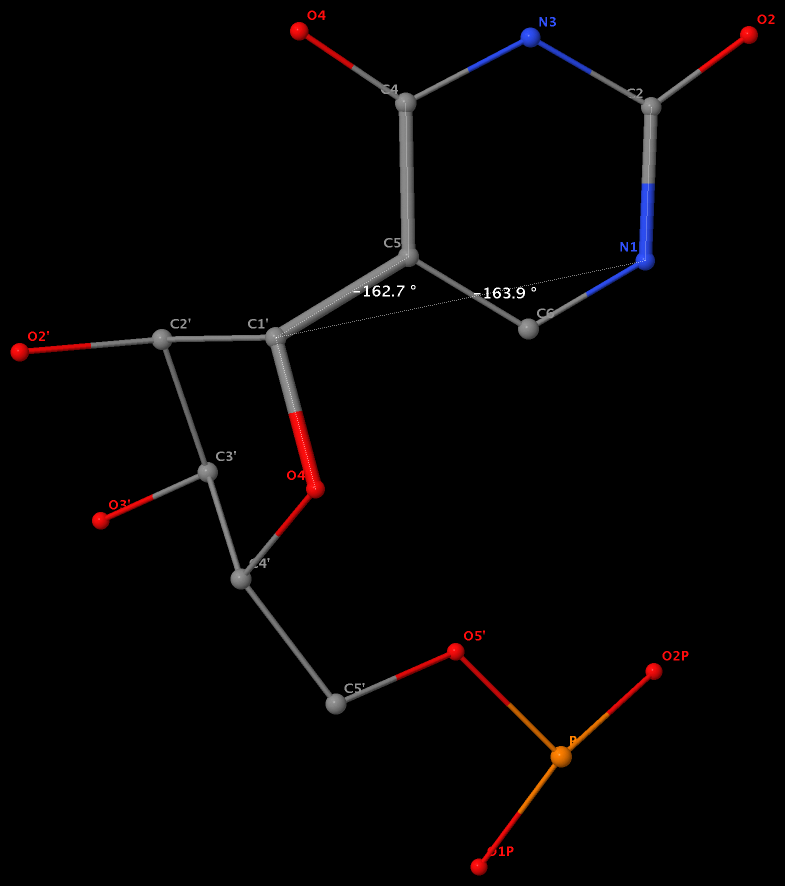
Pseudouridine (5-ribosyluracil, PSU) was the first identified modified nucleoside in RNA and is the most abundant. PSU is unique in that it has a C-glycosidic bond (C-C1′) instead of the N-glycosidic bond common to all other nucleosides, canonical or modified. It thus poses a problem as to how to calculate the χ torsion angle: should it be O4′-C1′-C5-C4, reflecting the actual glycosidic bond connection, or should the conventional definition O4′-C1′-N1-C2 still be applied literally? As a concrete example, the figure below shows the (slightly) different numerical values (–162.7° vs. –163.9°), as given by the two definitions, for PSU 6 on chain A of the PDB entry 3cgp (based on the 2009 Biochemistry article by Lin & Kielkopf titled X-ray structures of U2 snRNA-branchpoint duplexes containing conserved pseudouridines).
Needless to say, the specific definition of the χ torsion angle for PSU in RNA structures is a very subtle point, and I am not aware of any discussion on this issue in literature. In 3DNA, PSU is identified explicitly, and χ is defined by O4′-C1′-C5-C4. In NDB and a couple of other tools I am familiar with, χ for PSU is defined by O4′-C1′-N1-C2. Again using 3cgp (figure below) as an example, 3DNA gives –162.7°, whilst NDB gives –163.9°. Additionally, this distinction in N-C1′ vs. C-C1′ connection also comes into play when calculating the perpendicular distance from the 3′ phosphorus atom to the glycosidic bond, as per Richardson et al.

Except for pseudouridine, a nucleoside in DNA/RNA contains an N-glycosidic bond that connects the base to the sugar. The chi (χ) torsion angle, which characterizes the relative base/sugar orientation, is defined by O4′-C1′-N1-C2 for pyrimidines (C, T and U), and O4′-C1′-N9-C4 for purines (A and G).
Normally (as in A- and B-form DNA/RNA duplex), χ falls into the ranges of +90° to +180°; –90° to –180° (or 180° to 270°), corresponding to the anti conformation (Figure below, top). Occasionally, χ has values in the range of –90° to +90°, referring to the syn conformation (Figure below, bottom). Note that in left-handed Z-DNA with CG repeating sequence, the purine G is in syn conformation whilst the pyrimidine C is anti.
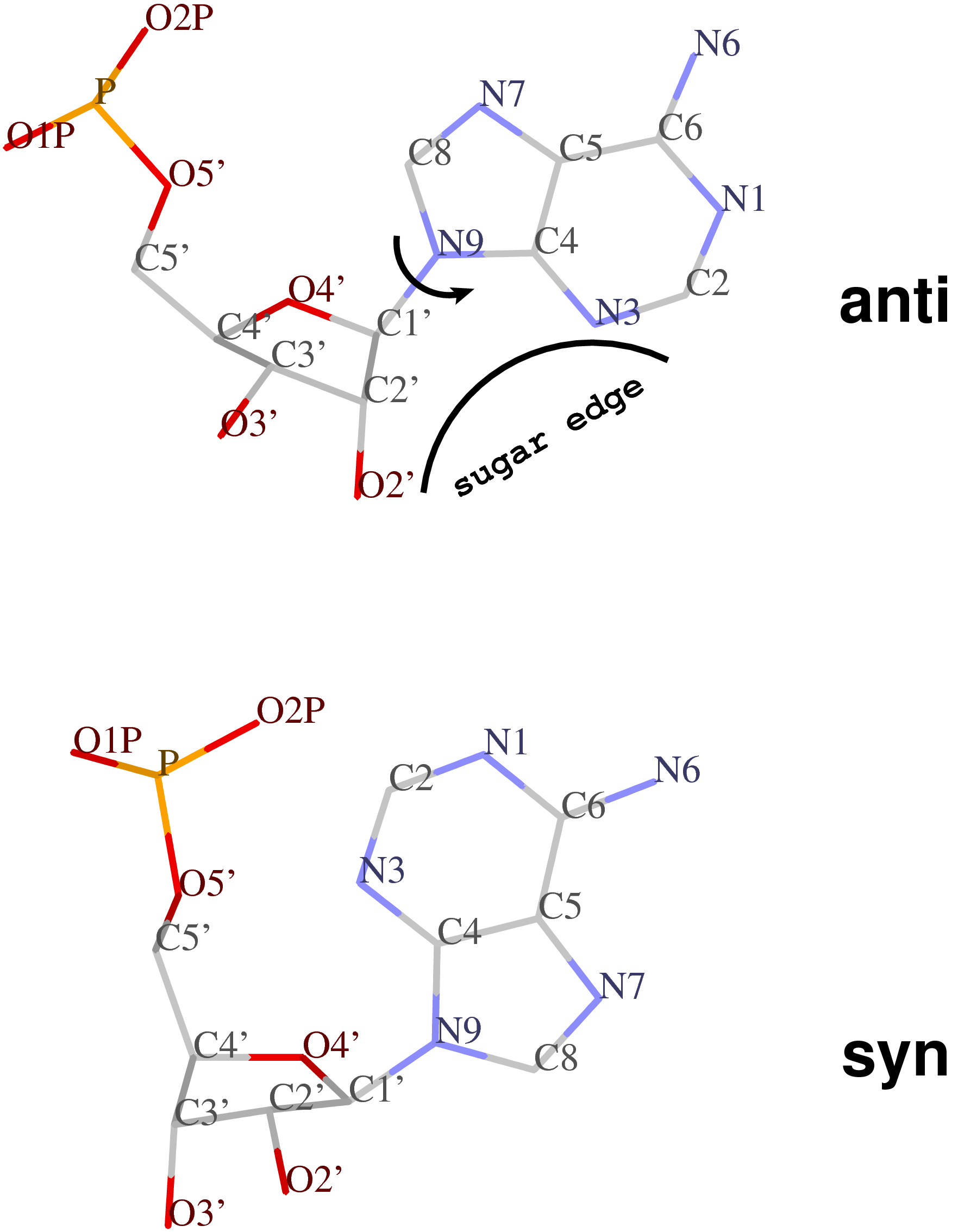
Presumably, the χ-related anti / syn conformation is a simple geometric concept. Nevertheless, the N-glycosidic bond and the corresponding χ torsion angle illustrate that the base and the sugar are two separate entities, i.e. there is an internal degree of freedom between them. In this respect, it is worth noting that the Leontis-Westhod sugar edge for base-pair classification corresponds to the anti form (as applied to RNA) only. When a base is flipped over into the syn conformation, the “sugar edge”, defined in connection with the minor (shallow) groove side of a nitrogenous bases, simply does not exist.
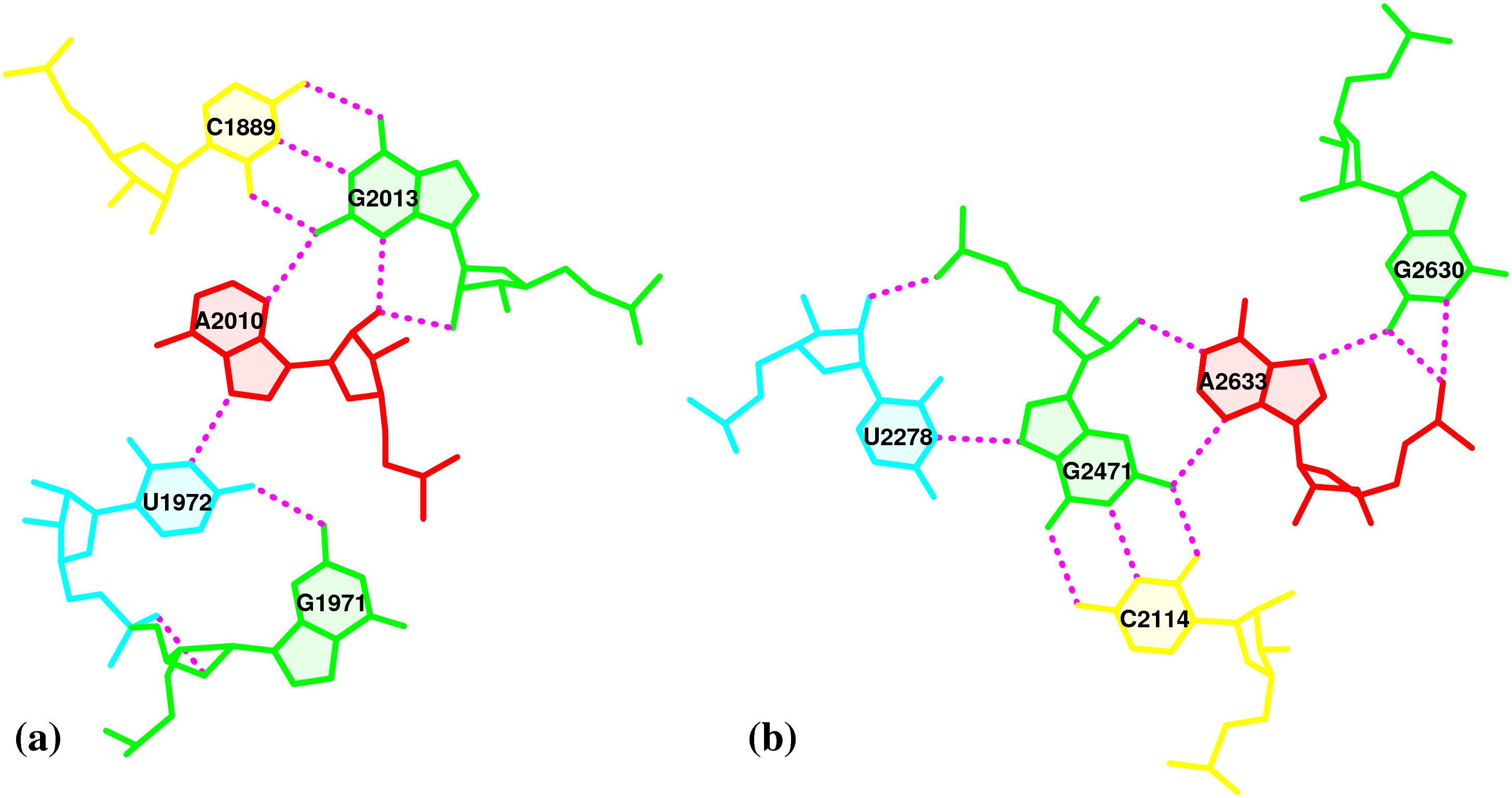
Base-flipping (anti / syn conformation switch) is one of the factors associated with the two possible relative orientations of the two bases in a pair, characterized explicitly in 3DNA as of type M+N or M–N since the 2003 NAR paper (Figure 2, linked below). I re-emphasized this distinction in our 2010 GpU dinucleotide platform paper (in particular, see supplementary Figure S2). Unfortunately, this subtle (but crucial, in my opinion) point has never been taken seriously (or at all) by the RNA community, even with 3DNA’s wide adoption. However, as people know 3DNA deeper/better and take RNA base-pair classification more rigorously, I have no doubt that the simplicity of this explicit distinction and the resultant full quantification of each and every possible base pair using standard geometric parameters will gradually be appreciated.

As of 3DNA v2.1, the output of the χ torsion angle is also associated with its classification in anti / syn conformation, among other new features (see for example the output for 6tna).

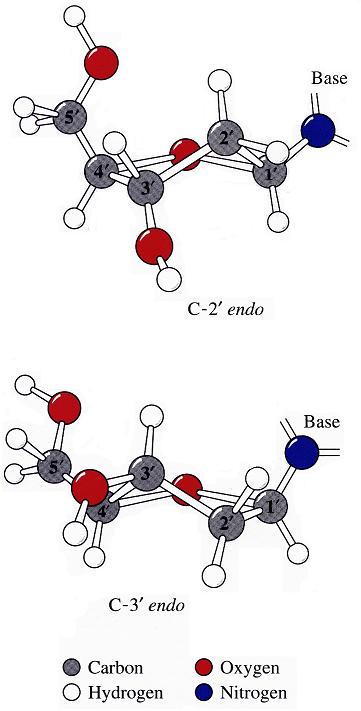
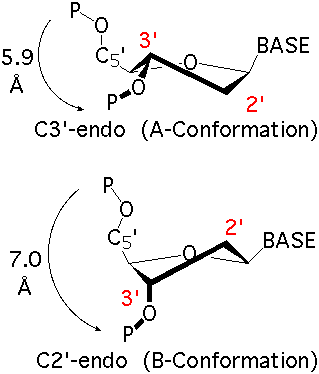
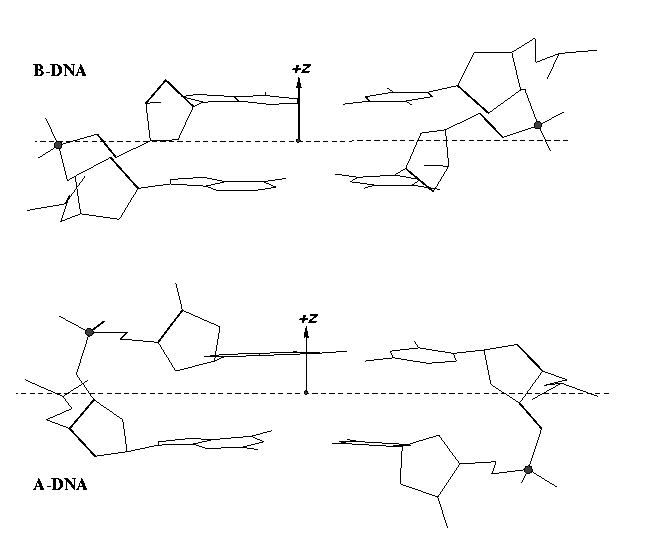
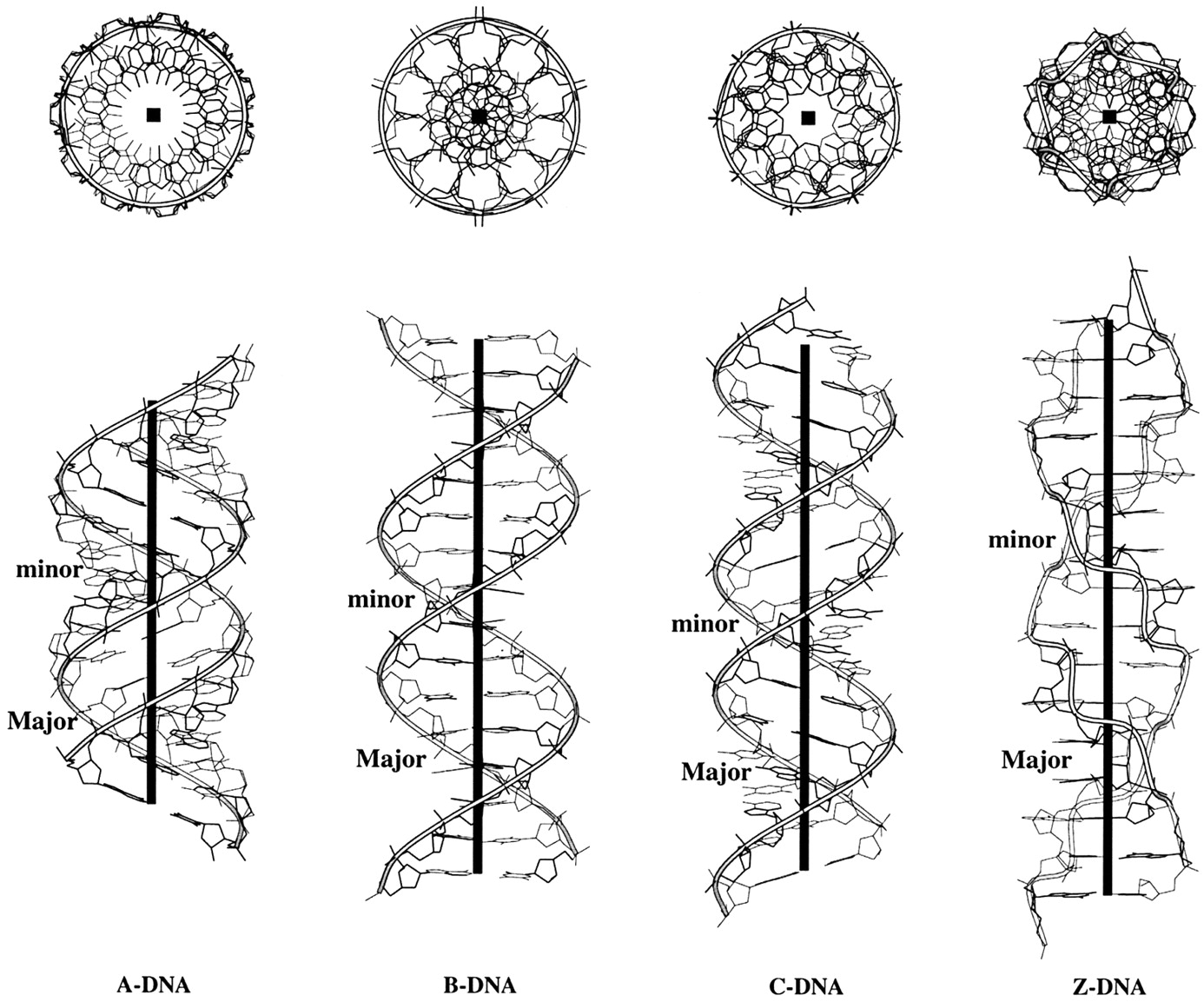
The sugar puckers in DNA/RNA structures are predominately in either C3′-endo (A-DNA or RNA) or C2′-endo (B-DNA; see Figure below, left), corresponding to the A- or B-form conformation in a duplex. In these two sugar conformations, the distance between neighboring phosphorus (P) atoms and the orientation of P relative to the sugar/bases are also dramatically different (figure below, right).
Recently, I carefully re-read some articles on RNA backbone conformation by Richardson et al., including:
I became intrigued by one of their observations: i.e., the correlation between the sugar pucker and a simple distance parameter:
C3′-endo and C2′-endo sugar puckers are highly correlated to the perpendicular distance between the C1′–N1/9 glycosidic bond vector and the following phosphate: > 2.9 Å for C3′-endo and < 2.9 Å for C2′-endo. (p.16 from the MolProbity paper).
Out of curiosity and for a better understanding of this correlation, I played around with some sample cases both visually and numerically. Overall, this involves a simple geometric calculation, i.e., the shortest distance from a point to a line in three-dimensional space. Given below is the Octave/Matlab script for calculating the distances for G175 and U176 of PDB entry 1jj2 (the large ribosomal subunit of Haloarcula marismortui):
function d = get_p3_nc_dist(P3, C1, N)
C1_N = N - C1; # vector from C1′ to N
nv_C1_N = C1_N / norm(C1_N); # normalized vector
C1_P3 = P3 - C1; # vector from C1′ to P3
proj = dot(C1_P3, nv_C1_N);
d = norm(C1_P3 - proj * nv_C1_N);
end
## G175
P3 = [70.104 112.366 44.586];
C1 = [73.017 109.666 45.304];
N9 = [74.445 109.380 45.288];
d1 = get_p3_nc_dist(P3, C1, N9) # 2.2 Å -- C2′-endo
## U176
P3 = [66.871 116.402 46.804];
C1 = [68.213 112.454 49.279];
N1 = [69.678 112.480 49.438];
d2 = get_p3_nc_dist(P3, C1, N1) # 4.6 Å -- C3′-endo
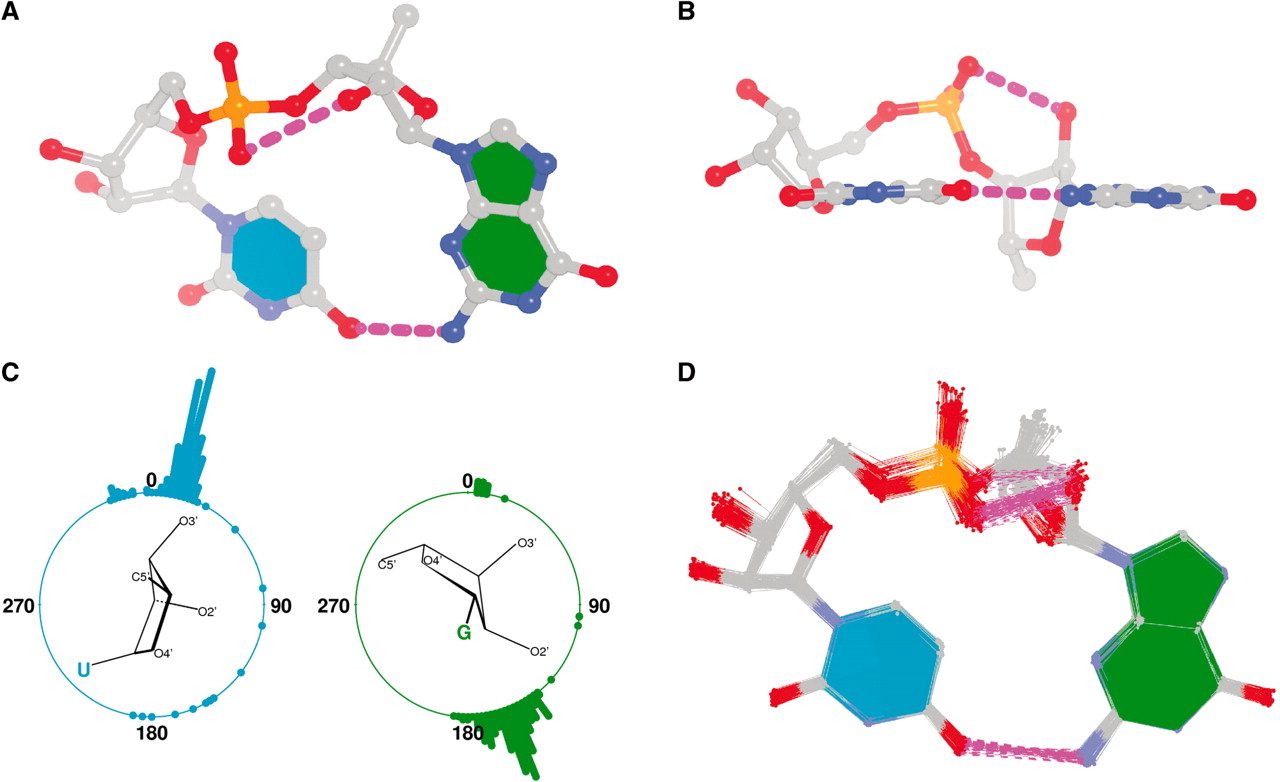
The GpU dinucleotide used in the above example forms a platform (see figure below), where the sugar of G175 adopts a C2′-endo conformation, and that of U176 C3′-endo. Indeed, the distance for G175 is 2.2 Å (< 2.9 Å); whilst the value for U176 is 4.6 Å (> 2.9 Å).

Note that the Richardson et al. articles focus on the RNA backbone, without paying attention to the base (pair) geometry. The 3DNA Zp parameter, which is the mean z-coordinate of the two P atoms in the mean reference frame of a dinucleotide step (see figure below), has been readily adapted to single-stranded RNA structures. For example, the vertical distances of the 3′ P atoms to the G175 and U176 base planes are 1.9 Å and 4.4 Å, respectively. Since base planes and the P atoms are the two most accurately located entities in a given nucleic acid structure, the nucleotide-based Zp variant is presumably more robust and discriminative than the distance from P to the glycosidic bond.
This new single-stranded based “Zp” parameter is available as of 3DNA v2.1.

RNA has three salient structural features (compared to DNA): it contains the ribose (not deoxyribose) sugar, it has the uracil (not thymine) base, and it is normally single (not double)-stranded. The O2′(G)…O2P(U) H-bond stabilized GpU dinucleotide platform may turn out to be the smallest unit with all those RNA hallmarks.
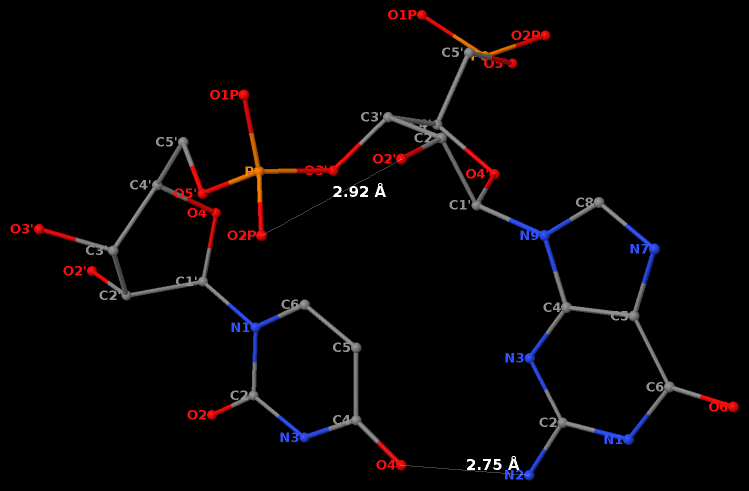
First, it must have the guanosine ribose to have the 2′-hydroxyl group form the O2′(G)…O2P(U) H-bond.
Second, the methyl group in position 5 of thymine would cause steric clash with guanosine, thus disrupting the N2(G)…O4(U) base-base H-bond to form the GpU dinucleotide platform.
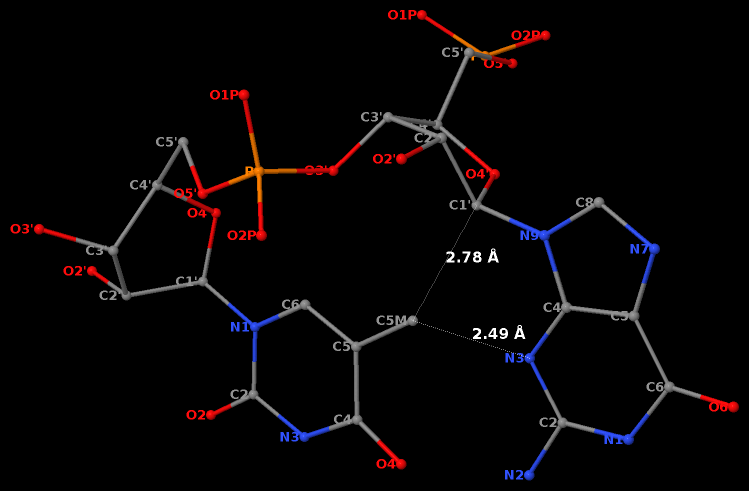
Third, a dinucleotide, by definition, is single-standed. The two H-bonds, plus the covalent linkage, makes the GpU platform extremely rigid (see Figure 1 of our 2010 NAR paper).
Moreover, the GpU platform is directional: swapping the two bases while keeping the sugar-phosphate backbone fixed does not allow for a base-base H-bond, thus no UpG dinucleotide platform.
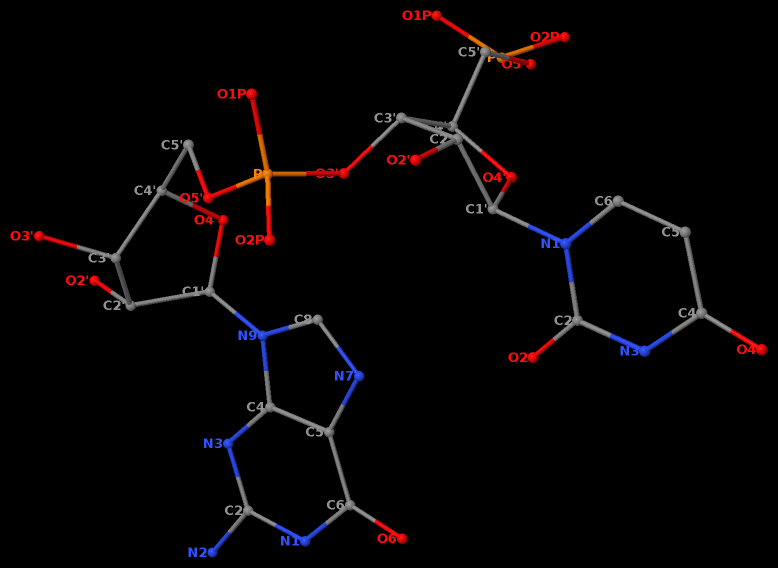
It worth noting that state-of-the-art quantum chemistry calculations have verified the importance of the O2′(G)…O2P(U) H-bond in stabilizing the GpU dinucleotide platform.

The least-squares (LS) fitting procedures presented below make use of well known mathematics. Indeed, the methods are so well known and widely used that it is somewhat difficult to locate the original references. In our previous effort to resolve the discrepancies among nucleic acid conformational analysis programs, we came across a variety of LS fitting procedures. Here we provide a detailed description, with step-by-step examples, of our implementation in 3DNA of two LS fitting algorithms based on a covariance matrix and its eigen-system. This post is the revised version of a note first made available in the “Technical Details” section of earlier 3DNA websites.
LS fitting between standard and experimental bases
Three analysis schemes — CompDNA, Curves/Curves+, and RNA — use LS procedures to fit a standard base with an embedded reference frame to an observed base structure. CompDNA and Curves/Curves+ take advantage of the conventional approach of McLachlan [“Least Squares Fitting of Two Structures.” J. Mol. Biol., 128, 74-79 (1979)], while the RNA program implements a closed-form solution of absolute orientation using unit quaternions first introduced by Horn. The two algorithms are mathematically equivalent for the most general cases, since the unit quaternion can be transformed to the rotation matrix given by McLachlan. The Horn method, however, is more straightforward and generally applicable; it can be applied even when one or both of the structures are perfectly planar, whereas the McLachlan approach fails.
Here we use the ideal adenine geometry derived from the high resolution crystal structures of model nucleosides, nucleotides, and bases. The x-, y-, and z-coordinates of the standard base, taken from the NDB, are listed below in the columns labeled sx, sy, and sz, respectively. s_(average) is the geometric center of the base.
sx sy sz
1 N9 0.213 0.660 1.287
2 C4 0.250 2.016 1.509
3 N3 0.016 2.995 0.619
4 C2 0.142 4.189 1.194
5 N1 0.451 4.493 2.459
6 C6 0.681 3.485 3.329
7 N6 0.990 3.787 4.592
8 C5 0.579 2.170 2.844
9 N7 0.747 0.934 3.454
10 C8 0.520 0.074 2.491
------------------------------------
s_(average): 0.4589 2.4803 2.3778
We similarly describe the coordinates of one of the adenine bases (the fifth nucleotide in the sequence strand) from the high resolution (1.4 Å) self-complementary d(CGCGAATTCGCG) dodecamer duplex determined by Williams and co-workers (PDB id: 355d). The experimental xyz coordinates are listed below in the columns labeled ex, ey, and ez. The geometric center is e_(average). Note that the atomic serial numbers from the PDB (first column) have been rearranged so that the atoms are in the same order as those of the ideal base listed above.
ex ey ez
91 N9 16.461 17.015 14.676
100 C4 15.775 18.188 14.459
99 N3 14.489 18.449 14.756
98 C2 14.171 19.699 14.406
97 N1 14.933 20.644 13.839
95 C6 16.223 20.352 13.555
96 N6 16.984 21.297 12.994
94 C5 16.683 19.056 13.875
93 N7 17.918 18.439 13.718
92 C8 17.734 17.239 14.207
------------------------------------
e_(average):16.1371 19.0378 14.0485
We collect the two sets of xyz coordinates in the 10 × 3 matrices S and E corresponding respectively to the standard and experimental bases. We then construct the 3 × 3 covariance matrix C between S and E using the following formula:
1 1
C = ------- [S' E - --- S' i i' E]
n - 1 n
=
0.2782 0.2139 -0.1601
-1.4028 1.9619 -0.2744
1.0443 0.9712 -0.6610
Here n, the number of atoms in each base, is 10, and i is an n x 1 column vector consisting of only ones. S' and i' are the transpose of matrix S and column vector i, respectively.
From the nine elements of the C matrix, we subsequently generate the 4 × 4 real symmetric matrix M using the expression:
| c11+c22+c33 c23-c32 c31-c13 c12-c21 |
M = | c23-c32 c11-c22-c33 c12+c21 c31+c13 |
| c31-c13 c12+c21 -c11+c22-c33 c23+c32 |
| c12-c21 c31+c13 c23+c32 -c11-c22+c33 |
=
1.5792 -1.2456 1.2044 1.6167
-1.2456 -1.0228 -1.1890 0.8842
1.2044 -1.1890 2.3447 0.6968
1.6167 0.8842 0.6968 -2.9011
The largest eigenvalue of matrix M is 4.0335, and its corresponding unit eigenvector is:
[ q0 q1 q2 q3 ] = [ 0.6135 -0.2878 0.7135 0.1780 ]
The rotation matrix R is deduced from the above eigenvector as below:
| q0q0+q1q1-q2q2-q3q3 2(q1q2-q0q3) 2(q1q3+q0q2) |
R = | 2(q2q1+q0q3) q0q0-q1q1+q2q2-q3q3 2(q2q3-q0q1) |
| 2(q3q1-q0q2) 2(q3q2+q0q1) q0q0-q1q1-q2q2+q3q3 |
=
-0.0817 -0.6291 0.7730
-0.1923 0.7710 0.6072
-0.9779 -0.0990 -0.1839
Following coordinate transformation with matrix R, the origin of the standard base is found to be displaced from the experimental structure by:
o = e_(average) - s_(average) R' = [15.8969 15.7701 15.1802]
The least-squares fitted coordinates (F) of the standard base atoms on the experimental structure are then given by:
F = S R' + i o
=
16.4592 17.0194 14.6699
15.7747 18.1925 14.4586
14.4899 18.4519 14.7542
14.1729 19.6974 14.4070
14.9343 20.6404 13.8420
16.2222 20.3472 13.5569
16.9832 21.2875 12.9925
16.6829 19.0585 13.8760
17.9183 18.4437 13.7219
17.7335 17.2396 14.2062
Here S is the (n x 3) matrix of original coordinates of the standard base, and as noted above, i is an n x 1 column vector consisting of only ones.
The difference matrix (D) between F and E, the (n x 3) matrix of original coordinates of the experimental base, and the root-mean-square (RMS) deviation between the two structures are found as:
D = E - F
=
0.0018 -0.0044 0.0061
0.0003 -0.0045 0.0004
-0.0009 -0.0029 0.0018
-0.0019 0.0016 -0.0010
-0.0013 0.0036 -0.0030
0.0008 0.0048 -0.0019
0.0008 0.0095 0.0015
0.0001 -0.0025 -0.0010
-0.0003 -0.0047 -0.0039
0.0005 -0.0006 0.0008
RMS deviation = 0.0054
It should be noted that if the standard base is already defined in terms of its reference frame, as in 3DNA (e.g., $X3DNA/config/Atomic_A.pdb), the vector o and the matrix R represent the best-fitted coordinate frame of the experimental base. Moreover, the three axes of the frame given by R are guaranteed to be orthonormal. If you want to get an insight of the LS fitting algorithm and a better understanding of how 3DNA derives its base reference frame, it’d be a valuable experience to repeat the above procedure with $X3DNA/config/Atomic_A.pdb.
Note: the algorithm does not apply to a molecule vs its inversion (an improper rotation) — thanks to Boris Averkiev for reporting this subtle point (see comments below). One possible remedy is to treat this edge case separately.
Base normal
Rather than fit a standard base to experimental coordinates, the CEHS, FREEHELIX, and NUPARM analyses perform a fitting of a LS plane to a set of atoms in order to define the base and base-pair normals. The covariance matrix based on the n x 3 matrix of experimental Cartesian coordinates E is diagonalized to find the vector normal to the best plane. Specifically, C is obtained using the above formula with S substituted by E. The normal vector then lies along the eigenvector that corresponds to the smallest eigenvalue. Note that the coefficient 1/(n-1) in the formula for calculating C has no effect on the direction of the eigenvectors but scales the magnitudes of the eigenvalues.
Using the above adenine base from the high resolution dodecamer duplex as an example, the covariance matrix C is:
C =
1.6680 -0.5015 -0.3253
-0.5015 2.0670 -0.5840
-0.3253 -0.5840 0.3061
The smallest eigenvalue of C, 8.26e-5, indicates that the base is almost perfectly planar. The corresponding unit eigenvector corresponding to the base normal is:
Base normal: 0.2737 0.3224 0.9062
Related topics:

As the old saying goes, a picture is worth a thousand words. To help you have a better idea of what 3DNA/DSSR is about, we’ve collected the following pictures; they serve to demonstrate selected features from 3DNA/DSSR’s versatile functionality.
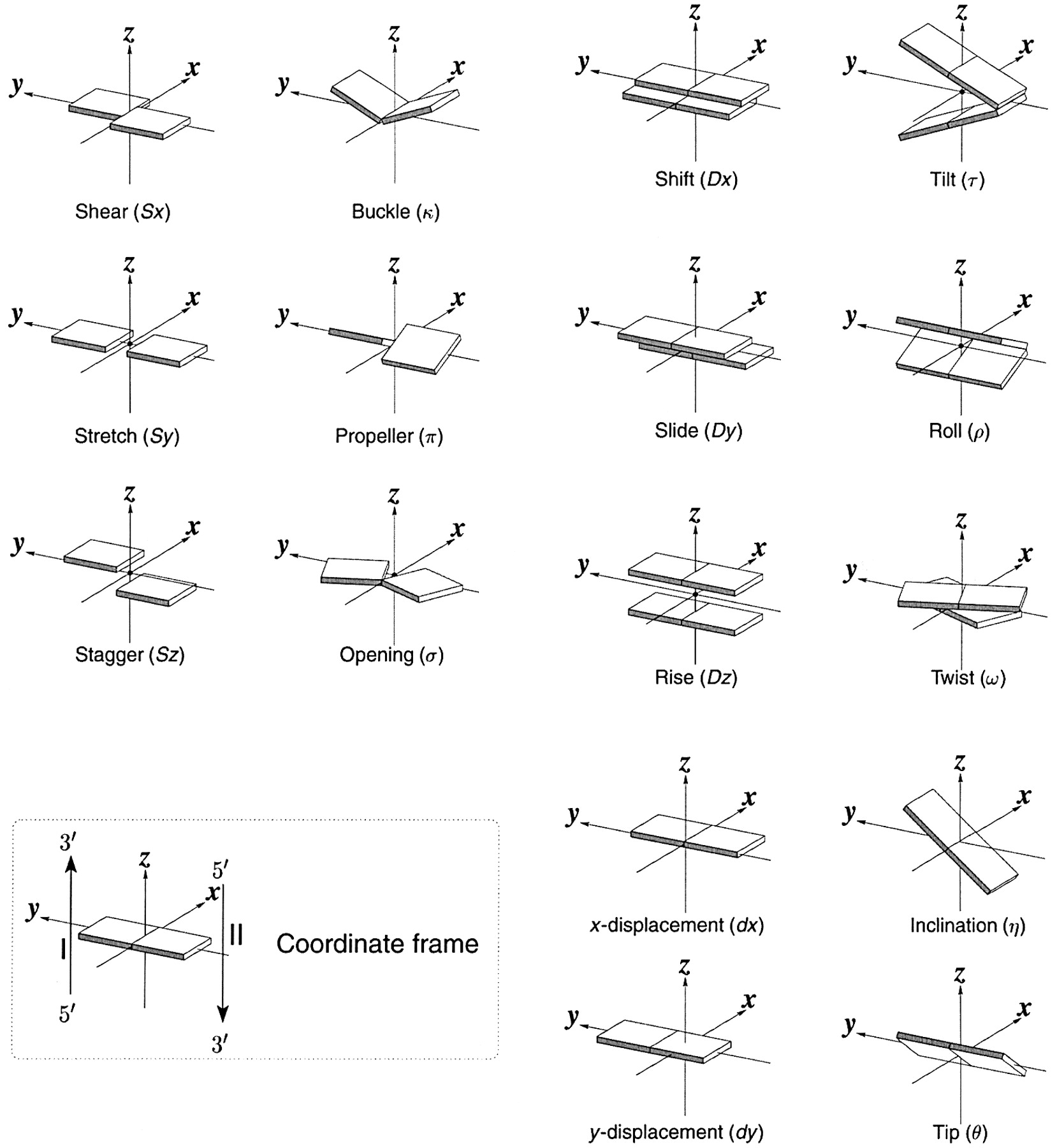
Schematic diagram of base-pair parameters
Influence of Slide and Roll on DNA helical conformation
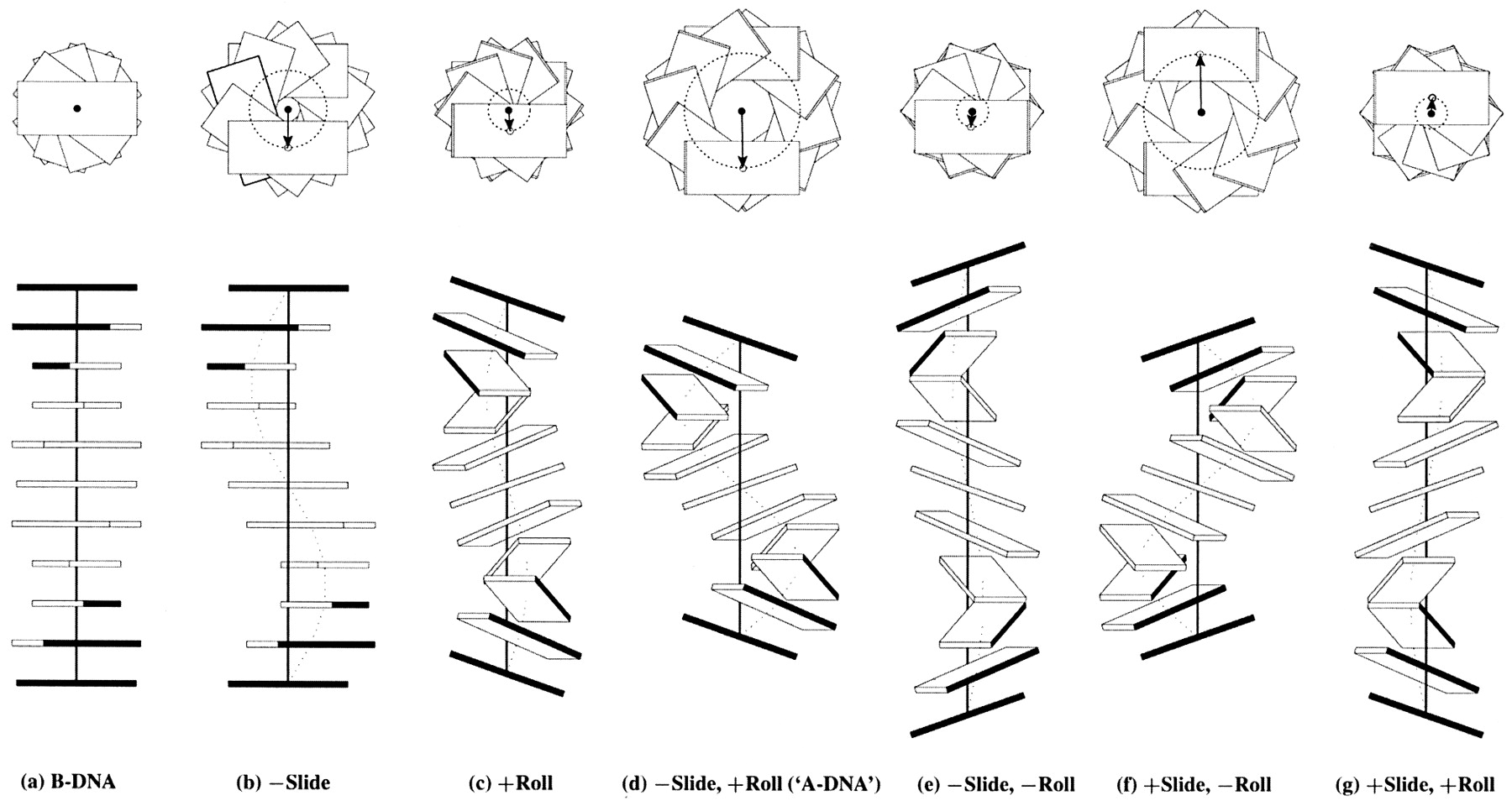
Roll-introduced DNA bending

Global bending of DNA associated with selective B → A conformational transformation

Canonical fiber models of A-, B-, C- and Z-DNA
3DNA-generated view of a four-way DNA–RNA junction (1egk)
3DNA-detected pentaplets in the large ribosomal subunit (1jj2)
Nucleic-acid-containing structures generated with w3DNA
Analysis of DNA with a B-Z junction (2acj, left) and detection of hydration patterns (right)
Schematics images auto-generated via blocview

Over the years, the fiber utility program has become a handy way to generate standard B-DNA and A-DNA structures, as evident from citations to 3DNA. Nevertheless, the currently collected 55 experimental fiber models, comprehensive as they are, do not include one for canonical double-stranded (ds) RNA or single-stranded (ss) RNA structures of generic A/C/G/U sequence.
This situation is best illustrated by a recent article by Charles Brooks and Hashim Al-Hashimi and their co-workers, titled Unraveling the structural complexity in a single-stranded RNA tail: implications for efficient ligand binding in the prequeuosine riboswitch [Nucleic Acids Research, 40(3) 1345–1355 (2012)] , where they wrote:
Idealized A-form structures were constructed using Insight II (Molecular Simulations, Inc.) correcting the propeller twist angles from +15° to –15° using an in-house program, as previously described (47). The complementary strand was removed and the resulting ssRNA used in NMR data analysis. B-form helices were constructed using W3DNA (48).
As of 3DNA v2.1, however, that’s no longer the case: now the fiber utility provides direct support for generating idealized dsRNA or ssRNA structures of arbitrary A/C/G/U sequence. As always, the new functionality can be best illustrated with examples. Let’s build ssRNAs of the wild-type (5’-AUAAAAAACUAA-3’) and A29C mutated form (5’-AUAACAAACUAA-3’) used in the work cited above:
fiber -r -s -seq=AUAAAAAACUAA wt-12nt.pdb
fiber -r -s -seq=AUAACAAACUAA mt-12nt.pdb
Here the -r option is for RNA, -s for a ss structure, and -seq for the specific base sequence. The generated ssRNA structure for the wild-type sequence is named wt-12nt.pdb, and that for the mutated sequence named mt-12nt.pdb.
Note that the new RNA model is based on Struther Arnott’s work of fiber A-DNA from calf thymus (#1 in the list). The dsRNA, as its dsDNA counterpart, has a helical twist of 32.7° and a helical rise of 2.548 Å. Relevant to the above citation, here the propeller twist angle of each base pair is –10.5°, a negative value similar to that observed in high-resolution x-ray crystal structures. Furthermore, you can easily verify the three numbers with the following commands:
fiber -r -seq=AUAAAAAACUAA wt-12nt.pdb
find_pair wt-12nt.pdb stdout | analyze stdin
In summary, it is very easy to generate canonical RNA structures with the revised fiber command. Through its integrated analysis routine, 3DNA can also be used to check structural features of the resultant RNA models. Moreover, as mentioned in the opening post What can 3DNA do for RNA structures? on the forum, 3DNA has much to offer in the filed of RNA structural bioinformatics.

At the C2B2 party this afternoon, I was asked the question: “Does 3DNA work for RNA?” Well, a good question, indeed. The short answer is definitely, YES. However, a detailed explanation is needed to address the underlying intuitive assumption: 3DNA is only for DNA.
- The name 3DNA was due to Dr. Olson, after we struggled quite a while. Initially, we played with NuStar (which was actually cited once by Richard Dickerson et al.), and Carnival etc. I still remember the day when Dr. Olson asked me “How about 3DNA?” We immediately reached an agreement: that’s it — what a cute name! Another advantage (as it becomes clear later): since 3DNA starts with ‘3’, it (mostly) shows up right at the top of many on-line lists of bioinformatics tools.
- Interpreted literally, 3DNA could mean 3-DNA, i.e., the three most common types of DNA: A-, B- and Z-form. That may be one of the reasons where the misconception that 3DNA is only for 3DNA comes from. Another reason could be that structural work on DNA is what the Olson lab best known for.
- The number ‘3’ in 3DNA should also be associated with its three key components: analysis, rebuilding and visualization. In a sense, this is my favorite.
- Of course, 3DNA stands for 3D-NA, 3-Dimensional Nucleic Acids, as expressed explicitly in the titles of our two 3DNA papers (2003 NAR and 2008 NP).
The applications of 3DNA to RNA structures can be broadly categorized as follows:
- Automatically detect all existing base-pairs, Watson-Crick (A-U, G-C, wobble G-U) or non-canonical, using a set of simple geometric criteria. Furthermore, it has a unique base-pair classification system based on the six numerical structural parameters, suitable for database storage and search.
- Automatically detect all triplets or higher-order base-associations.
- Automatically detect double helical regions, regardless of backbone connection, thus ideal for finding pseudo-continuous coaxial stacking.
- The above three features are seamlessly integrated with the visualization component to allow for easy generation of publication quality images. See the 3DNA 2008 NP paper for detailed examples.
As further examples, the following two RNA publications take advantage of find_pair from 3DNA:
It is well worth noting that the base-pair detecting algorithm in RNAView is based on an earlier version of find_pair, a basic fact ignored in the RNAView publication.
In summary, 3DNA works for RNA as well as for DNA, and more.