I recently noticed a bioRxiv preprint, titled Role of RNA Guanine Quadruplexes in Favoring the Dimerization of SARS Unique Domain in Coronaviruses by a European team consisting of scientists from France, Italy, and Spain. The abstract is as follows. Figure 1 shows a schematic representation of the mRNA with a G-Quadruplex structure, functioning in a healthy cell and an infected cell by coronavirus.
Coronaviruses may produce severe acute respiratory syndrome (SARS). As a matter of fact, a new SARS-type virus, SARS-CoV-2, is responsible of a global pandemic in 2020 with unprecedented sanitary and economic consequences for most countries. In the present contribution we study, by all-atom equilibrium and enhanced sampling molecular dynamics simulations, the interaction between the SARS Unique Domain and RNA guanine quadruplexes, a process involved in eluding the defensive response of the host thus favoring viral infection of human cells. The results obtained evidence two stable binding modes with guanine quadruplexes, driven either by electrostatic (dimeric mode) or by dispersion (monomeric mode) interactions, are proposed being the dimeric mode the preferred one, according to the analysis of the corresponding free energy surfaces. The effect of these binding modes in stabilizing the protein dimer was also assessed, being related to its biological role in assisting SARS viruses to bypass the host protective response. This work also constitutes a first step of the possible rational design of efficient therapeutic agents aiming at perturbing the interaction between SARS Unique Domain and guanine quadruplexes, hence enhancing the host defenses against the virus.
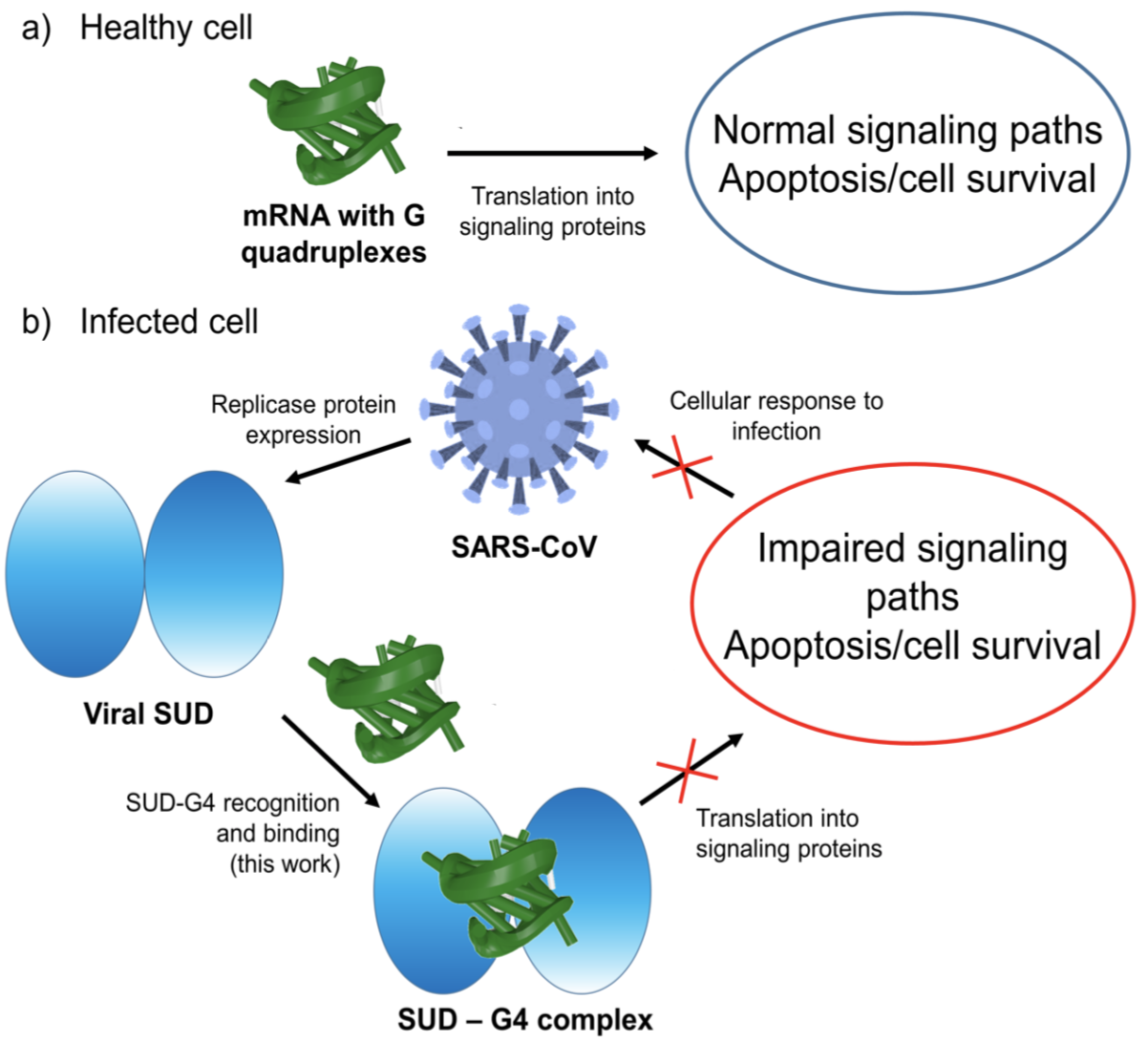
Figure 1) Schematic representation of the mRNA function in a) a healthy cell and b) an infected cell by coronavirus. Panel b) showcases the influence of viral SUD binding to G4 sequences of mRNA that encodes crucial proteins for the apoptosis/cell survival regulation and other signaling paths.
In the manuscript, the software tools employed in this MD study are described as below:
… Both protein and RNA have been described with the amber force field including the bsc1 corrections, and the MD simulations have been performed in the constant pressure and temperature ensemble (NPT) at 300K and 1 atm. All MD simulations have been performed using the NAMD code and analyzed via VMD, the G4 structure has also been analyzed with the 3DNA suite.
I am glad that 3DNA has played a role in the analysis of G-quadruplexes in this timely contribution. In particular, I would like to draw attention of the community to 3DNA-DSSR which has a brand-new module dedicated to the automatic identification and comprehensive characterization of G-quadruplexes. The DSSR-annotated G-quadruplexes from the PDB should be of great interest to a wide audience, especially the experimentalists. As a concrete example, the authors noted that “The crystal structure … of the oligonucleotide (pdb 1J8G) have been chosen coherently with the experimental work performed by Tan et al”. Follow the link to see results of DSSR-derived G-quadruplex features in PDB entry 1J8G and you are guaranteed to see features not available elsewhere.
Note added on July 9, 2020: This paper has been published in J. Phys. Chem. Lett. 2020, 11, 5661−5667.
