Recently I was surprised by some cases of nucleotides with missing atoms in PDB entry 1pns. The story started like this: 3DNA/DSSR maps various nucleotide names to one-letter codes, based on the data file baselist.dat (see post Modified nucleotides in the PDB). In the meantime, 3DNA/DSSR internally assigns a nucleotide as either purine or pyrimidine, by virtue of coordinates of base atoms. Be definition, purines should only include A/a/G/g/I/i, and pyrimidines C/c/T/t/U/u/P/p. However, no consistency check has been implemented in DSSR until just now.
I first noticed the inconsistency between residue name and atom coordinates for nucleotide A6 on chain U (hereafter referred to as U.A6) in 1pns. The nucleotide has standard name ‘ A’, obviously a purine. However, somehow DSSR classified it as a pyrimidine based on atomic coordinates. Upon further check of the PDB data file, I found the following remarks:
REMARK 470 MISSING ATOM REMARK 470 THE FOLLOWING RESIDUES HAVE MISSING ATOMS(M=MODEL NUMBER; REMARK 470 RES=RESIDUE NAME; C=CHAIN IDENTIFIER; SSEQ=SEQUENCE NUMBER; REMARK 470 I=INSERTION CODE): REMARK 470 M RES CSSEQI ATOMS REMARK 470 A U 6 N9 C8 N7 REMARK 470 G U 8 N9 C8 N7 REMARK 470 A U 12 N9 C8 N7 REMARK 470 A U 13 N9 C8 N7 REMARK 470 A U 14 N9 C8 N7
The atomic coordinates for U.A6 are as below:
ATOM 34447 P A U 6 81.861 37.210 78.651 1.00378.87 P ATOM 34448 OP1 A U 6 80.631 37.121 77.831 1.00378.87 O ATOM 34449 OP2 A U 6 81.665 37.221 80.119 1.00378.87 O ATOM 34450 O5' A U 6 82.707 38.495 78.212 1.00378.87 O ATOM 34451 C5' A U 6 83.948 38.777 78.887 1.00378.87 C ATOM 34452 C4' A U 6 84.600 40.000 78.276 1.00378.87 C ATOM 34453 O4' A U 6 84.975 39.698 76.901 1.00378.87 O ATOM 34454 C3' A U 6 83.714 41.239 78.153 1.00378.87 C ATOM 34455 O3' A U 6 83.654 41.968 79.369 1.00378.87 O ATOM 34456 C2' A U 6 84.403 42.015 77.020 1.00378.87 C ATOM 34457 O2' A U 6 85.564 42.655 77.474 1.00378.87 O ATOM 34458 C1' A U 6 84.834 40.864 76.105 1.00378.87 C ATOM 34459 C5 A U 6 82.033 39.296 74.209 1.00378.87 C ATOM 34460 C6 A U 6 82.941 39.553 75.166 1.00378.87 C ATOM 34461 N6 A U 6 81.170 39.949 72.090 1.00378.87 N ATOM 34462 N1 A U 6 83.830 40.588 75.041 1.00378.87 N ATOM 34463 C2 A U 6 83.843 41.410 73.939 1.00378.87 C ATOM 34464 N3 A U 6 82.899 41.124 72.974 1.00378.87 N ATOM 34465 C4 A U 6 81.968 40.108 73.016 1.00378.87 C
No atom records for N7, C8 and N9. So far, so good. However, surprise came when I visualized U.A6 in Jmol, as shown in the following image. Note here atom N1 is connected to C1’ as in pyrimidines, and N6 is bonded to C4!
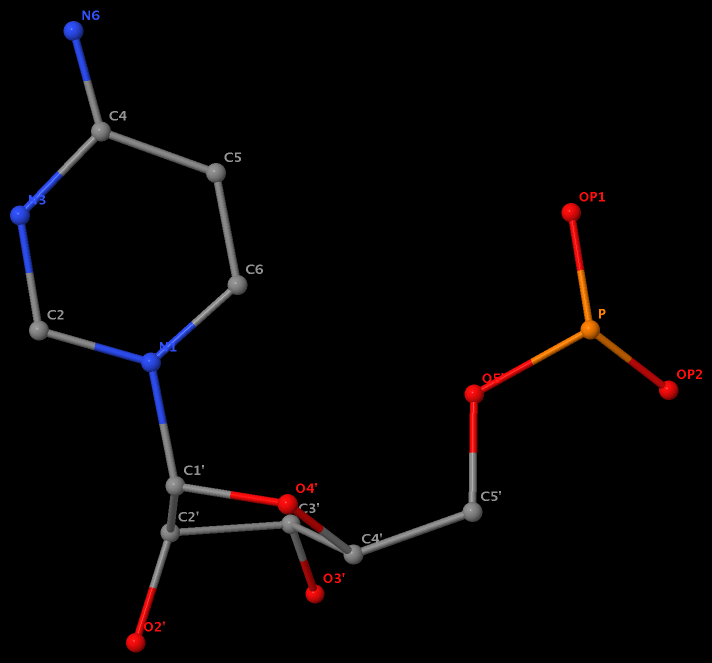
The same issue also exists for U.G8 (see figure below), U.A12, U.A13, and U.A14.
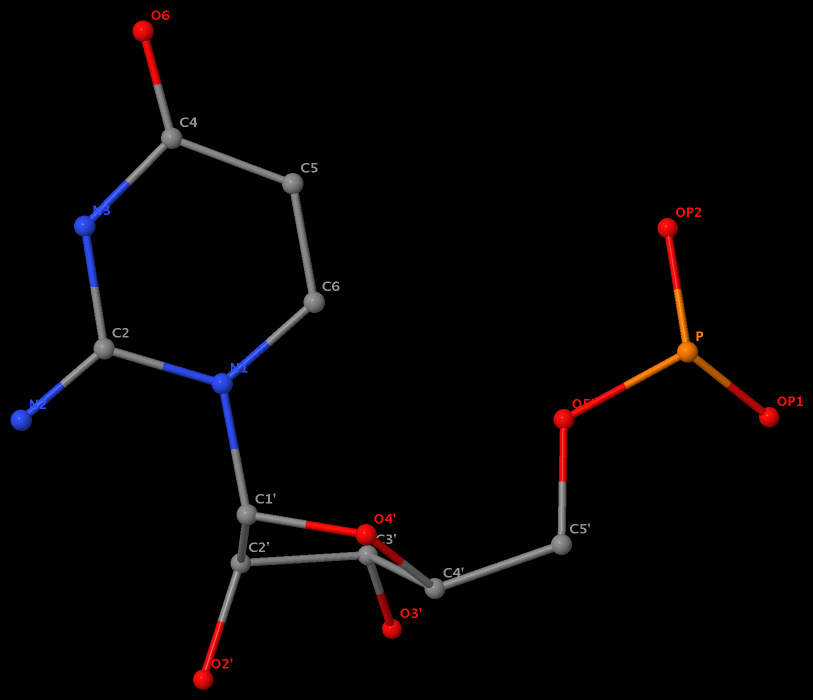
It is beyond my imagination to understand why such weird cases exist in the PDB, even given the lousy resolution (8.7 Å) of 1pns.
