A G-quadruplex (G4) is composed of stacks of G-tetrad where four guanines form four G•G pairs in a circular, planar fashion. Specifically, the G•G pairs of the G-tetrad (see Fig. 1A below) in G4 are of type M+N according to 3DNA/DSSR: i.e., G+G with the local z-axes of pairing guanines in parallel. Moreover, the G+G pair is uniquely quantified by three base-pair parameters: Shear, Stretch, and Opening with mean values [+1.6 Å, +3.5 Å, –90º] or [–1.6 Å, –3.5 Å, +90º], corresponding to the cWH (cW+M) or cHW (cM+W) types of LW (DSSR) classifications, respectively. This pair is numbered VI in the list of 28 base pairs with two or more H-bonds between base atoms, compiled by Saenger.
In addition to the standard G-tetrad configuration as normally seen in G4 structures, a so-called pseudo-G-tetrad form (see Fig. 1B below) is reported in a 2013 paper titled Duplex-quadruplex motifs in a peculiar structural organization cooperatively contribute to thrombin binding of a DNA aptamer. (PDB entry 4i7y). In a 2017 publication from the same group, Through-bond effects in the ternary complexes of thrombin sandwiched by two DNA aptamers, another form of pseudo-G-tetrad (Fig. 1C) is found in PDB entries 5ew1 and 5ew2.
Clearly, pseudo-G-tetrads are very different from the normal G-tetrad, in terms of base pairing patterns. The G-tetrad is highly regular with the same type of G+G pairs, with the O6 atoms pointing to the middle of the circle. The two pseudo-G-tetrads are less regular, and they differ from each other as well, by flipping G12 from syn (Fig. 1B) to trans (Fig. 1C).
These distinctions stand out even more by filling the up-face (+z-axis outwards) of a guanine base in green while leaving the down-face (+z-axis inwards) empty (G5 in Fig. 1B, G5 and G12 in Fig. 1C). So in G-tetrad (Fig. 1A), all four guanines have their positive z-axis point towards the viewer, corresponding to all four G+G pairs. In one pseudo-G-tetrad (Fig. 1B), G5 has its positive z-axis pointing away from the viewer. So G5–G7 and G5–G16 pairs are of the M–N type. The other type of pseudo-G-tetrad (Fig. 1C) has the opposite orientation for G12. Finally, Fig. 1D shows schematically PDB entry 4i7y where the G-tetrad and a pseudo-G-tetrad are directly stacked, creating a two-layered pseudo-G-quadruplex.
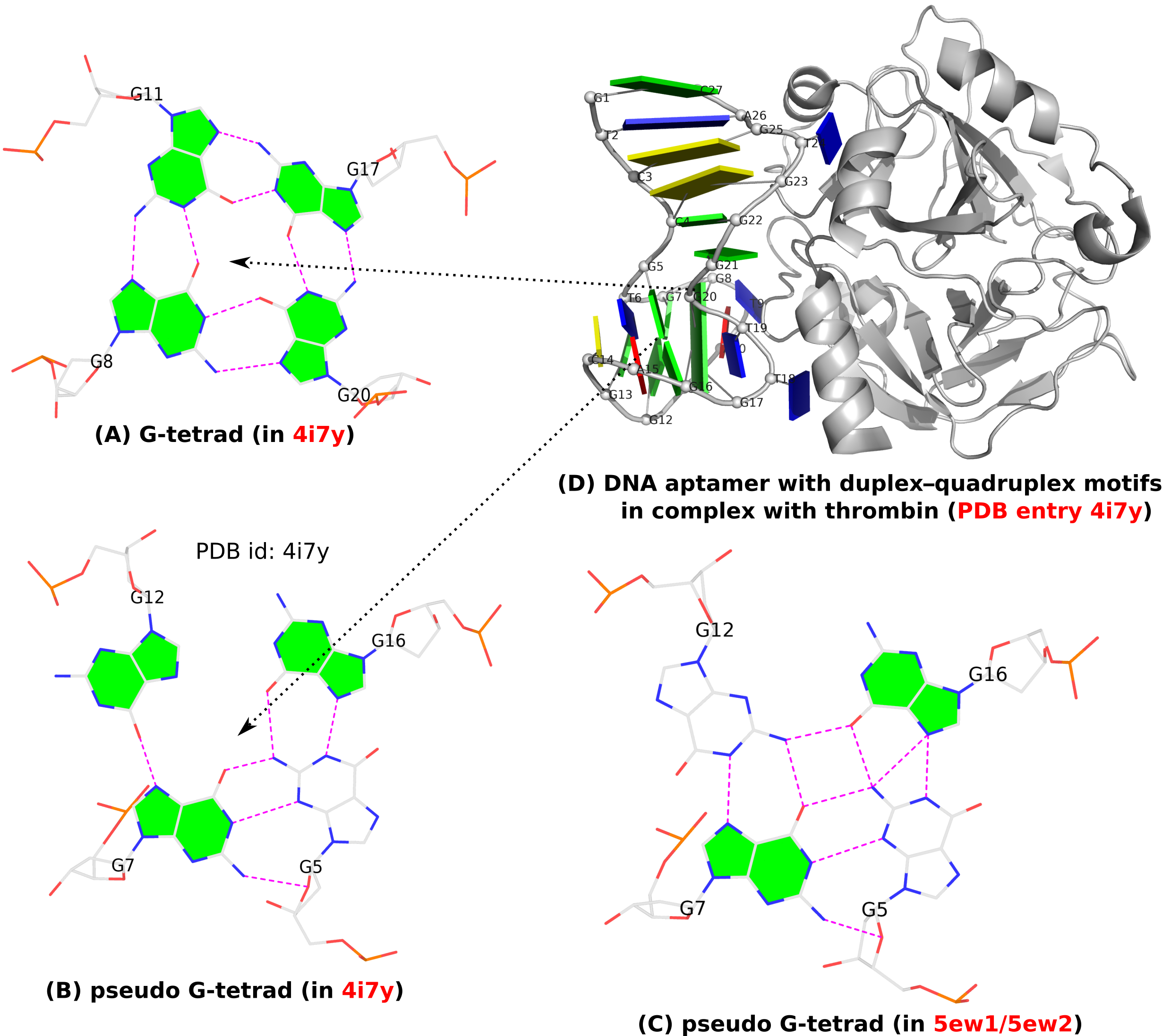
Figure 1. (A) G-tetrad, (B-C) two types of pseudo-G-tetrads, and (D) the complex of a DNA-apatmer with thrombin. G-tetrads were automatically identified by 3DNA-DSSR. The images were created using DSSR and PyMOL.
