In late March, I was approached by Mike May. He was then writing an article for Biocompare about DNA-protein interactions and asked me to answer a few questions on “What features of 3DNA be used in studying DNA-protein interactions?” and “Please provide 1-2 examples.” Initially, I was a bit surprised by the contact. Thus, I visited his online profile and Amazon Author Page. I also read a couple of his previous publications. Impressed by his track records, I answered his requests and our following communications were as smooth and professional as I could have ever imagined.
The paper The Best Ways to Study DNA and Protein Interactions has now been published, and is freely accessible. It includes the following content:
3DNA creator and maintainer Xiang-Jun Lu mentioned a couple of ways that the software has been used. For example, he noted that “3DNA can analyze all DNA-protein complexes in the Protein Data Bank—PDB—in an automatic, consistent, and robust manner,” and other bioinformatic resources have adopted this feature of 3DNA. He added that scientists have used 3DNA to “understand the structural basis on how transcription factors recognize methylated DNA.” Moreover, 3DNA is continuously developed. A new feature of 3DNA is the automatic identification and comprehensive characterization of G-quadruplexes, a noncanonical DNA structure formed from guanine-rich base sequences.
The bioinformatics resource I used as an example is the paper DNAproDB: an expanded database and web-based tool for structural analysis of DNA–protein complexes by the Rohs lab. The phrase “to understand the structural basis on how transcription factors recognize methylated DNA” refers to the article Toward a mechanistic understanding of DNA methylation readout by transcription factors by the Bussemaker lab. Both works employed DSSR and SNAP, two sophisticated programs I created and maintained over the past ten years, and they have largely obsoleted the original 3DNA suite of programs.
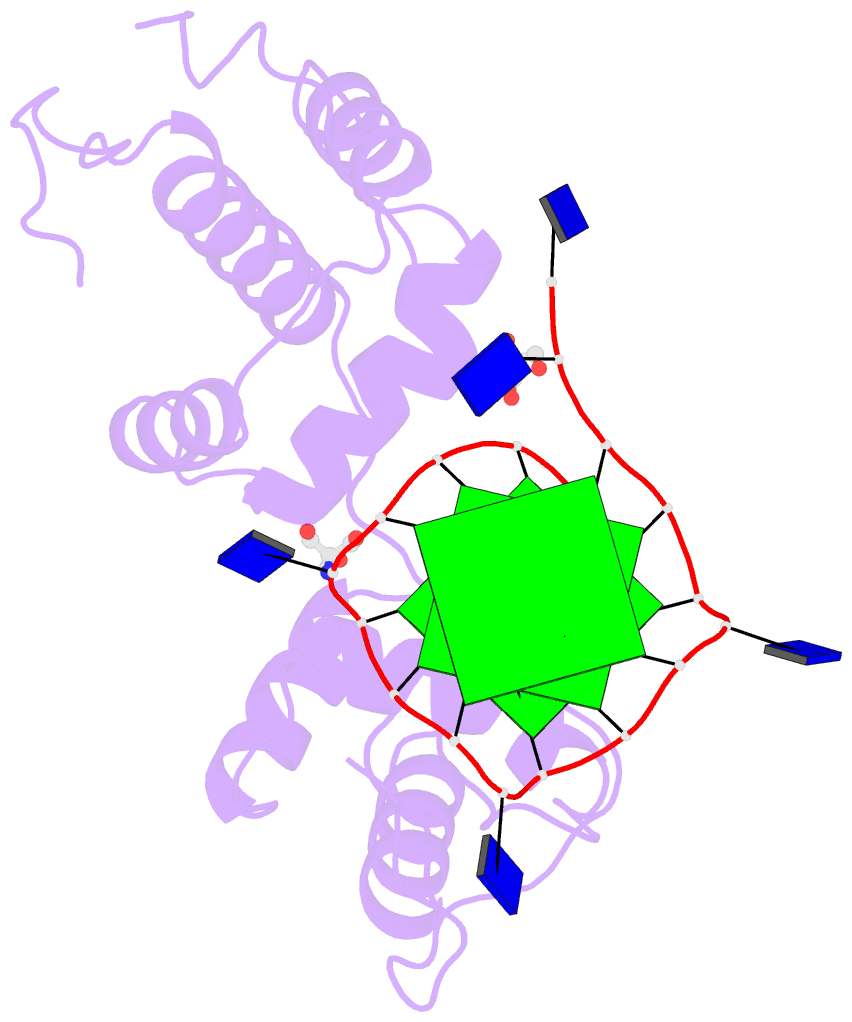
The image I provided is a DSSR-PyMOL schematic based on PDB entry 6LDM. The 6LMD picture features a G-quadruplex, for which DSSR comes with an unmatched set of features (including automatic identification and comprehensive annotations). See the http://g4.x3dna.org/ page for survey results, curated using DSSR, of all G-quadruplexes from the PDB.
This image of a protein-DNA complex (PDB entry 6LDM) shows the protein (purple), the DNA G-quadruplex (green) and thymine (blue). The image was created using the 3DNA-DSSR program and PyMOL. Image courtesy of Xiang-Jun Lu.