Canonical bases (A, C, G, T and U) in nucleic acid structures have standard atom names, shown below using the Watson-Crick A–T and G–C pairs. Ring atoms of adenine, for example, are named (N1, C2, N3, C4, C5, C6, N7, C8, N9) respectively.
Four characters are reserved for atom names in the PDB format. The convention, as seen in files downloaded from the RCSB PDB, is to put the two-character base name in the middle, as in .N1.. Note that here each dot (.) is used for a space character to make it stand out.
Long time ago, I became aware a PDB format variant where the base name is left-aligned, as in N1... This case has ever since been properly handled by 3DNA (including DSSR and SNAP). While checking submitted entries to web-DSSR, I recently noticed yet another PDB format variation in labeling base names with the format of ..N1 (i.e., right-aligned). Without taking this special variant of PDB format into consideration, 3DNA/DSSR reported that “no nucleotides found!” Once the issue is known, however, fixing it is straightforward. As of May 4, 2015, 3DNA v2.2, DSSR and SNAP can all handle this special PDB variant correctly.
Over the years, I have come across many PDB variants claimed to compliant with the loosely defined format. If you find 3DNA or DSSR is not working as expected, it is likely the coordinate file in the self-claimed ‘PDB format’ is at fault. Wherever practical, I’ve tried to incorporate as many non-standard variants as possible.

The NDB (Nucleic Acid Database) is a valuable resource dedicated to “information about experimentally-determined nucleic acids and complex assemblies.” Over the years, however, I’ve gradually switched from NDB to PDB (Protein Data Bank) for my research on nucleic acid structures. NDB is derived from PDB and presumably should contain all nucleic acid structures available in the PDB. However, at the time of this writing (on April 9, 2015), the NDB says: “As of 8-Apr-2015 number of released structures: 7430” and the PDB states “7611 Nucleic Acid Containing Structures”. So PDB has 7611-7430=181 more entries of nucleic acid structures than the NDB, possibly due to a lag in NDB’s processing of newly released PDB structures. Another issue is the inconsistency of the NDB identifier: early entries have e.g. bdl084 for B-DNA (355d in PDB), but now NDB seems to use the same id as the PDB (e.g., 4p5j).
The RCSB PDB maintains a weekly-updated, summary file named pdb_entry_type.txt in pure text format (check here for a list of useful summary files), containing “List of all PDB entries, identification of each as a protein, nucleic acid, or protein-nucleic acid complex and whether the structure was determined by diffraction or NMR.” An excerpt of the file is shown below:
108m prot diffraction
109d nuc diffraction
109l prot diffraction
109m prot diffraction
10gs prot diffraction
10mh prot-nuc diffraction
110d nuc diffraction
110l prot diffraction
.................................
102m prot diffraction
103d nuc NMR
Specifically, a nucleic acid structure contains the (sub)string nuc in the second field, where prot-nuc means a protein-RNA/DNA complex. This text file is trivial to parse, and the atomic coordinates files (in PDB or PDBx/mmCIF format) for all nucleic acid structures can be automatically downloaded from the RCSB PDB using a script.
It is worth noting that DSSR is checked against all nucleic acid structures in the PDB at the time of each release to ensure that it does not crash. I update my local copy of nucleic acid structures each week, and run DSSR on the new entries. This process not only provides me an opportunity to keep pace with new developments in the field but also allows me to keep refining DSSR as needs arise.

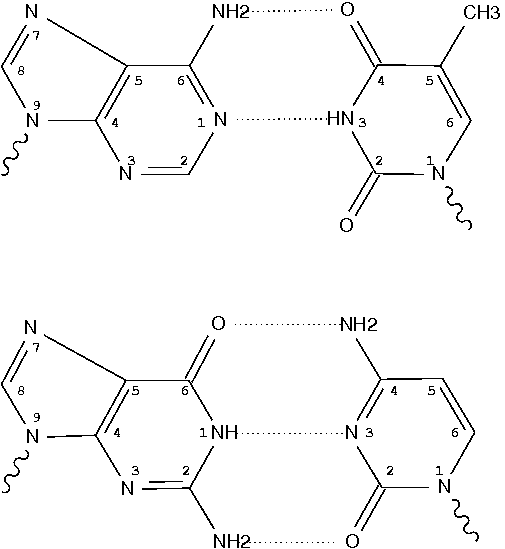
Pseudouridine (5-ribosyluracil, PSU) is the most abundant modified nucleotide in RNA. It is unique in that it has a C-glycosidic bond (C-C1′) instead of the N-glycosidic bond (N-C’) common to all other nucleotides, canonical or modified. In 3DNA, the one-letter code for PSU is assigned to the upper case ‘P’, reserving the lower case ‘p’ for its modified variants. Distinguishing PSU from standard U (or T) is important for deriving sensible base-pair parameters and the χ torsion angle.
 |
 |
|
| PSU |
3TD |
Recently, I came across 3TD (see figure above) in PDB entry 5afi. 3TD is a modified variant of PSU, with a methyl group attached to N3. In 3DNA v2.1 v2.1-2015mar11, 3TD is abbreviated to ‘p’ to signify its connection to PSU.
In the list of recognized nucleotides (‘baselist.dat’) distributed with 3DNA, there are two other residues mapped to ‘p’: FHU and P2U (see figure below). As is often the case, it is the chemical structure, not the 3-letter PDB ligand identifier (or even full chemical name), that shows clearly to what 3DNA 1-letter abbreviation a residue matches.
 |
 |
|
| FHU |
P2U |

A single-stranded RNA molecule can fold back onto itself to form various loops delineated by double helical stems, as shown in the figure below [taken from the Nearest Neighbor Database website from the Turner group].
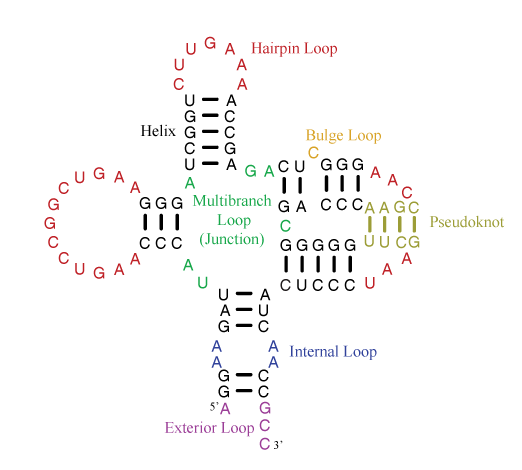
Of special note is the exterior loop (at the bottom) which includes the 5′ and 3′ ends of the sequence. The Mfold User Manual defines the exterior loop as such:
The collection of bases and base pairs not accessible from any base pair is called the exterior (or external) loop … . It is worth noting that if we imagine adding a 0th and an (n + 1)st base to the RNA, and a base pair 0.(n+1), then the exterior loop becomes the loop closed by this imaginary base pair. … The exterior loop exists only in linear RNA.
While each of the other loops (hairpin, bulge, internal or junction) forms a closed ‘circle’ with two neighboring bases connected by either a canonical pair or backbone covalent bond, the ‘exterior loop’ has only an imaginary pair to close the 5′ and 3′ ends of the sequence. Moreover, the two ends of an RNA molecule are not necessarily close in three-dimenional space, as may be implied in the above secondary structure diagram. For example, in the H-type pseudoknotted structure 1ymo from human telomerase RNA, the 5′ and 3′ ends are on the opposite sides.
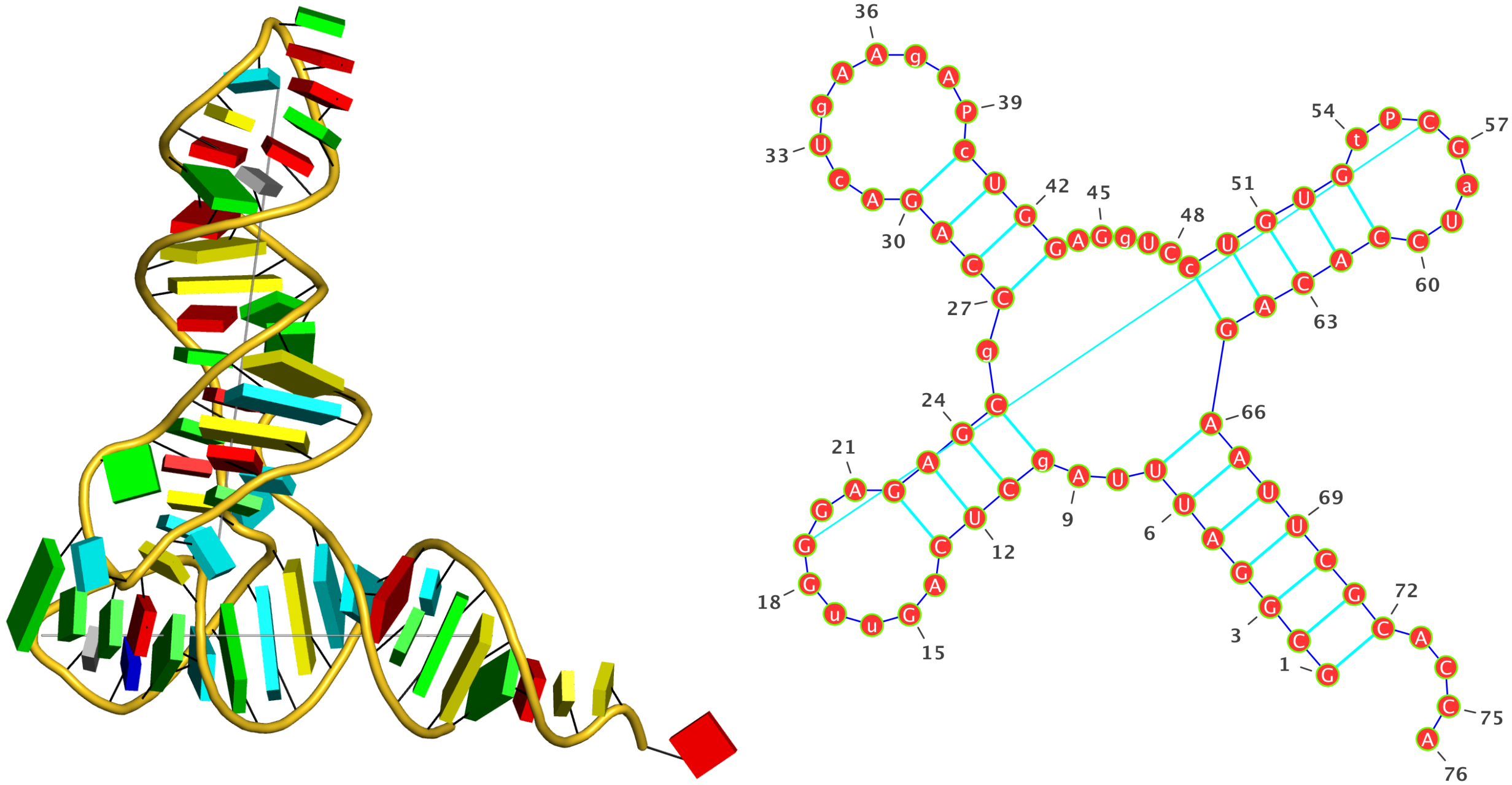
DSSR does not has the concept of an ‘exterior loop’ due to its lack of a closing pair to form a ‘circle’. Instead, each of the 5′ and 3′ dangling ends is taken as a ‘non-loop single-stranded segment’, if applicable. For the crystal structure of yeast phenylalanine tRNA (1ehz, see the figure at the bottom), the relevant portion of DSSR output is as below. Note that since the 5′ end is paired, only the single-stranded region at the 3′ end is listed. Presumably, the ‘exterior loop’ in this case would also include the G1—C72 pair, with the imaginary closing pair connecting G1 and A76.
List of 1 non-loop single-stranded segment
1 nts=4 ACCA A.A73,A.C74,A.C75,A.A76

Dot bracket notation (dbn) is a popular format to represent RNA secondary structures. Initially introduced by the ViennaRNA package, dbn uses dots (.) for unpaired bases, and matched parentheses () for the canonical Watson-Crick A-T and G-C or the wobble G-U pairs. This compact representation was designed for fully nested (i.e., pseudoknot free) RNA secondary structures in a single RNA molecule. Over the years, it has been extended to cover pseudoknots (of possibly higher orders) using matched pairs of [], {}, and <> etc.
To derive dbn from three-dimensional atomic coordinates with DSSR, I was faced with an issue on how to represent multiple RNA chains (molecules). A closely related yet practical problem is chain breaks, as in x-ray crystal structures where disordered regions may not have fitted coordinates. I searched but failed to find any ‘standard’ way to account for chain breaks or multiple molecules in dbn. The commonly used programs for visualizing RNA secondary structure diagrams that I tested at that time did not take such cases into consideration — they simply showed all bases as if they were from a single continous RNA chain.
I discussed the issue with Dr. Yann Ponty, the maintainer of the popular VARNA program. After a few around of email exchanges, we introduced an extra symbol (&) in both sequence and dbn to designate multiple chains or breaks within a chain to communicate between DSSR and VARNA.
As an example, the DSSR-derived dbn for the double-stranded DNA structure 355d (the famous Dickerson dodecamer) is as below:
Secondary structures in dot-bracket notation (dbn) as a whole and per chain
>355d nts=24 [whole]
CGCGAATTCGCG&CGCGAATTCGCG
((((((((((((&))))))))))))
>355d-A #1 nts=12 [chain] DNA
CGCGAATTCGCG
((((((((((((
>355d-B #2 nts=12 [chain] DNA
CGCGAATTCGCG
))))))))))))
As another example, the PDB entry 2fk6 contains a tRNA with chain breaks — nucleotides 26 to 45 are missing from the structure (see figure below). The DSSR-derived dbn is as follows — note the * at the end of the header line.
>2fk6-R #1 nts=53 [chain] RNA*
GCUUCCAUAGCUCAGCAGGUAGAGC&GUCAGCGGUUCGAGCCCGCUUGGAAGCU
(((((((..((((.....[..))))&...(((((..]....)))))))))))).
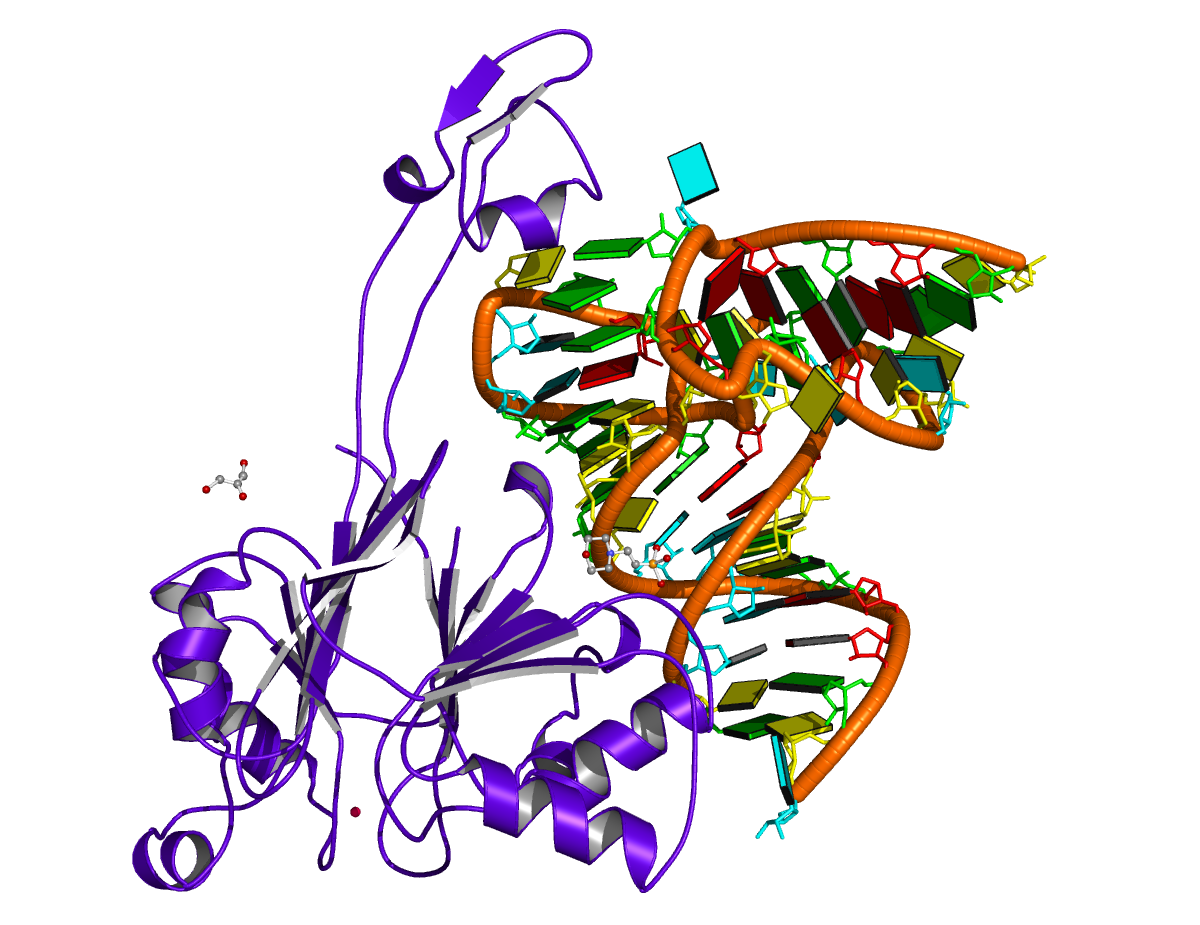
It is worth mentioning a subtle point in DSSR-derived dbn with multiple chains, i.e., the order of the chains may make a difference! The point is best illustrated with a concrete example — here, 4un3, the crystal structure of Cas9 bound to PAM-containing DNA target. Based on the data file downloaded directly from the PDB (4un3.pdb), the relevant portions of DSSR output are:
****************************************************************************
Special notes:
o Cross-paired segments in separate chains, be *careful* with .dbn
****************************************************************************
This structure contains *1-order pseudoknot
o You may want to run DSSR again with the '--nested' option which removes
pseudoknots to get a fully nested secondary structure representation.
o The DSSR-derived dbn may be problematic (see notes above).
****************************************************************************
Secondary structures in dot-bracket notation (dbn) as a whole and per chain
>4un3 nts=120 [whole]
AUAACUCAAUUUGUAAAAAAGUUUUAGAGCUAGAAAUAGCAAGUUAAAAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUG&CAATACCATTTTTTACAAATTGAGTTAT&AAATGGTATTG
((((((((((((((((((((((((((..((((....))))....))))))..(((..).)).......((((....)))).&[[[[[[[[))))))))))))))))))))&...]]]]]]]]
>4un3-A #1 nts=81 [chain] RNA
AUAACUCAAUUUGUAAAAAAGUUUUAGAGCUAGAAAUAGCAAGUUAAAAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUG
((((((((((((((((((((((((((..((((....))))....))))))..(((..).)).......((((....)))).
>4un3-C #2 nts=28 [chain] DNA
CAATACCATTTTTTACAAATTGAGTTAT
[[[[[[[[))))))))))))))))))))
>4un3-D #3 nts=11 [chain] DNA
AAATGGTATTG
...]]]]]]]]
The notes in the DSSR output is worth paying attention to. Specifically, it reports a “*1-order pseudoknot” — note also the *! Here the target DNA chain C comes before DNA chain D in the PDB file. The 5′-end bases in chain C pair with bases in D, and the 3′-end bases in C pair with RNA bases in chain A. There exist pairs crossing along the ‘linear’ sequence position-wise, hence the reported “pseudoknot”. However, simply reverse DNA chains C and D, i.e., moving chain D before C (in file 4un3-ADC.pdb), the “pseudoknot” will be gone, as shown below:
****************************************************************************
Secondary structures in dot-bracket notation (dbn) as a whole and per chain
>4un3-ADC nts=120 [whole]
AUAACUCAAUUUGUAAAAAAGUUUUAGAGCUAGAAAUAGCAAGUUAAAAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUG&AAATGGTATTG&CAATACCATTTTTTACAAATTGAGTTAT
((((((((((((((((((((((((((..((((....))))....))))))..(((..).)).......((((....)))).&...((((((((&))))))))))))))))))))))))))))
>4un3-ADC-A #1 nts=81 [chain] RNA
AUAACUCAAUUUGUAAAAAAGUUUUAGAGCUAGAAAUAGCAAGUUAAAAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUG
((((((((((((((((((((((((((..((((....))))....))))))..(((..).)).......((((....)))).
>4un3-ADC-D #2 nts=11 [chain] DNA
AAATGGTATTG
...((((((((
>4un3-ADC-C #3 nts=28 [chain] DNA
CAATACCATTTTTTACAAATTGAGTTAT
))))))))))))))))))))))))))))
Notes added on March 19, 2015
- It has drawn to my attention that the NUPACK uses ‘+’ instead of ‘&’ as the symbol to separate multiple chains (or chain breaks). In fact, DSSR has an undocumented option
--dbn_break which can be set to any of the character in the string &.:,|+. The ‘&’ symbol was chosen for communication with VARNA which requires ‘&’, at least up to now. This is an excellent example showing the efforts that I have put into the little details while developing DSSR.
- The issue on proper ordering of multiple chains to avoid crossing lines (false pseudoknots) has been formally addressed by Dirks et al. in their 2007 article titled Thermodynamic analysis of interacting nucleic acid strands (SIAM Rev, 49, 65-88), specifically in Section 2.1 (Fig. 2.1). Applying that algorithm to nucleic acid structures, however, is beyond the scope of DSSR. The program strictly respects the ordering of chains and nucleotides within a given PDB or PDBx/mmCIF file, but outputs warning messages where necessary to draw users’ attention. As another example, I’ve recently noticed that DNA duplexes produced by Maestro (a product of Schrödinger) list nucleotides of the complementary strand in 3′ to 5′ order to match the 5′ to 3′ directionality of the leading strand for each Watson-Crick pair (See below).
****************************************************************************
Special notes:
o nucleotides out of order
****************************************************************************
Secondary structures in dot-bracket notation (dbn) as a whole and per chain
>ga62_ca62_1m_in nts=24 [whole]
GGCGAATTCCGG&C&C&G&C&T&T&A&A&G&G&C&C
((((((((((((&)&)&)&)&)&)&)&)&)&)&)&)
>ga62_ca62_1m_in-1-A #1 nts=12 [chain] DNA
GGCGAATTCCGG
((((((((((((
>ga62_ca62_1m_in-1-B #2 nts=12 [chain] DNA
C&C&G&C&T&T&A&A&G&G&C&C
)&)&)&)&)&)&)&)&)&)&)&)

I’m going to attend the Biophysical Society (BPS) 59th Annual Meeting to be held during February 7-11 at Baltimore, Maryland. In last year’s BPS annual meeting (San Francisco, California), I was delighted to come across a few 3DNA users at poster sessions. I thought this post may help to connect me with some DSSR/3DNA users in the coming meeting.
Want to have a meetup at Baltimore? Please drop me a message!

Recently I came across the ligand thiamine pyrophosphate (TPP) in some RNA riboswitch structures. I was a bit surprised by the atom names adopted for the ligand by the PDB. See figures below for the chemical structure of TPP from the RCSB PDB website (first), and the three-dimensional structure of the ligand from the riboswitch 2gdi (second).

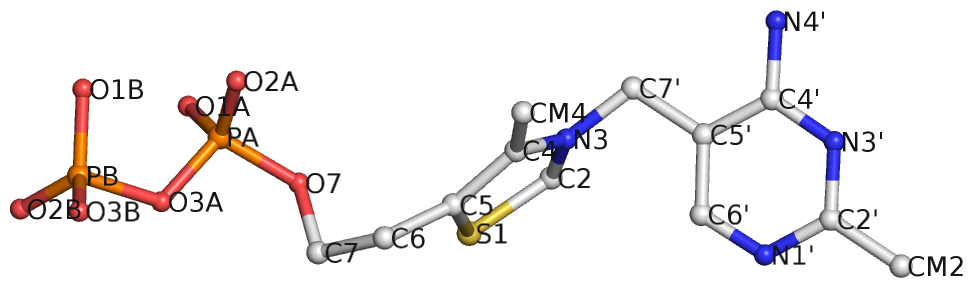
Specifically, the planar base-like moiety at the right has atom names ending with prime. To my knowledge, only sugar atom names of DNA and RNA nucleotides have the prime suffix, such as the 2′-hydroxyl group in RNA.
The RCSB webpage for TPP shows that currently there are 107 entries in the PDB, among which 100 are from proteins, 6 from RNA, and one in a RNA-protein complex. It is not clear to me whether the prime-bearing names in TPP are following any documented ‘standard’ or convention. DSSR is nevertheless taking a note of such ‘weird’ cases.

As of today, the number of registered users on the 3DNA Forum has reached 2000. Over the past three years, the annual average of resignations is 650, corresponding to approximately 1.8 per day. While many registrations use free email services (gmail, hotmail or yahoo, etc), a significant portion (especially more recent ones) employs their job email (e.g., .edu). This is clear sign of increasing trust the community puts in the Forum.
To ensure the 3DNA Forum spam-free, I’ve adhered a zero-tolerance policy of any trolling or suspicion activities. The anti-spam software has played a big role in making this clean status feasible, as is evident from the note: “120,933 Spammers blocked up until today”.
From a scientific perspective, all posted questions have been addressed promptly, normally within hours. Instead of feeling like a burden, maintaining the Forum and answering user questions have been a pleasure. I’d love to see more questions or posts on the Forum.

With great pleasure, I read the following annoancement from Rajendra Kumar on the 3DNA Forum:
Re: do_x3dna: a tool to analyze DNA/RNA in molecular dynamics trajectories
« Reply #1 on: Today at 10:53:31 AM »
Hello,
I have now made a new website for do_x3dna
(http://rjdkmr.github.io/do_x3dna). This website contains detailed
documentation for do_x3dna program and Python APIs.
Documentation for Python API is now available
(http://rjdkmr.github.io/do_x3dna/apidoc.html).
Few tutorials about the Python APIs are also now available
(http://rjdkmr.github.io/do_x3dna/tutorial.html).
Thanks.
With best regards,
Rajendra
Browsing through the do_x3dna website, I am impressed by the extensive documentation and tutorial. Clearly, do_x3dna has pushed the boundaries (in applicability and documentation) of the x3dna_ensemble Ruby script distributed with 3DNA v2.1.
As noted in GitHub page, do_x3dna has been developed to analyze fluctuations in DNA or RNA structures in molecular dynamics (MD) trajectories. It can be used for GROMACS MD trajectories, as well as those from NAMD and AMBER. It leaves no doubt that do_x3dna will boost 3DNA’s applications in the increasingly active field of DNA/RNA MD simulations.











