A recent thread on the 3DNA Forum discussed 'Rebuilding Z-DNA' by extending an existing structure. The 3DNA rebuild program allows users to generate DNA or RNA structures with any user-specific sequence and corresponding base-pair/step parameters. This process is rigorous for atomic coordinates of base (and C1') atoms: running analyze on the rebuilt structure will yield the same set of parameters that users initially input. For more details, see the 2003 3DNA paper, the 2015 DSSR paper, and the DSSR User Manual.
The challenge lies in modeling the backbones. For right-handed A- or B-form DNA, users can build full-atomic models with canonical backbone conformations of C3'-endo or C2’-endo sugar conformations and anti glycosidic bonds. However, left-handed Z-DNA has unique structural features—such as syn-G, CpG, and GpC dinucleotides as building blocks instead of single nucleotides—that are not fully addressed by the 3DNA rebuild program.
DSSR (Pro version or the Academic v2.5.2) offers a solution by providing tools to build extended Z-DNA structures with proper backbones. The commands are as follows:
x3dna-dssr -i=1qbj.pdb1 --select-chains='D E' --delete-water -o=model.pdb
x3dna-dssr tasks -i=model.pdb --frame-pair=last -o=model1-ref-last.pdb
# poly d(GC) : poly d(GC)
x3dna-dssr fiber --z-dna --repeat=1 -o=conn.pdb
x3dna-dssr tasks -i=conn.pdb --frame-pair=first --remove-pair -o=ref-conn.pdb
x3dna-dssr tasks --merge-file='model1-ref-last.pdb ref-conn.pdb' -o=temp1.pdb
x3dna-dssr tasks -i=temp1.pdb --frame-pair=last --remove-pair -o=temp2.pdb
x3dna-dssr tasks -i=model.pdb --frame-pair=first -o=model1-ref-first.pdb
x3dna-dssr tasks --merge-file='temp2.pdb model1-ref-first.pdb' -o=duplicate-model.pdb
x3dna-dssr --order-residue -i=duplicate-model.pdb -o=temp3.pdb --po-bond=3.6
x3dna-dssr --renumber-residue -i=temp3.pdb -o=temp4.pdb
x3dna-dssr --connect-file -i=temp4.pdb -o=1qbj-duplicate.pdb --po-bond=3.6
The logic behind these commands is very straightforward, but technical details may look a bit complex for the uninitiated:
- The first command extracts the Z-DNA duplex consisting of chains D and E from PDB entry
1qbj.pdb1 (the first biological unit) and remove water molecules (model.pdb). The Z-DNA duplex has sequence: CGCGCG/CGCGCG.
- The next command sets the Z-DNA duplex (
model.pdb) into the reference frame of the last base pair, i.e., G-C (model1-ref-last.pdb).
- The
fiber model consists of the GpC dinucleotide step (conn.pdb), which is then set into the reference frame of the first base pair (G-C). The first G-C pair is removed from the coordinate file ref-conn.pdb which consists of only one C-G pair.
- The two PDB files,
model1-ref-last.pdb and ref-conn.pdb, share a common reference frame and are merged into a single PDB file (temp1.pdb).
- The merged PDB file (
temp1.pdb) is then set into the reference frame of last base pair(i.e., C-G) which is removed from the resulting coordinate file (temp2.pdb). Now the job of the GpC fiber connector is done.
- The Z-DNA duplex (
model.pdb) is once again set into the reference frame of the first base pair (i.e., C-G), leading to the coordinate file model1-ref-first.pdb.
- The two PDB files,
temp2.pdb and model1-ref-first.pdb, both consist of the same Z-DNA duplex but are in different orientations. They now share a common reference frame and are merged into the extended Z-DNA duplex (1qbj-duplicate.pdb).
- The last three commands (with options
--order-residue, --renumber-residue, --connect-file) are bookkeeping steps to ensure proper order and numbering of nucleotides along each chain, and generate the CONECT record for smooth view in PyMOL.
The final PDB coordinate file (1qbj-duplicate.pdb) can be downloaded, and visualized in DSSR-enabled cartoon-block representation as below:
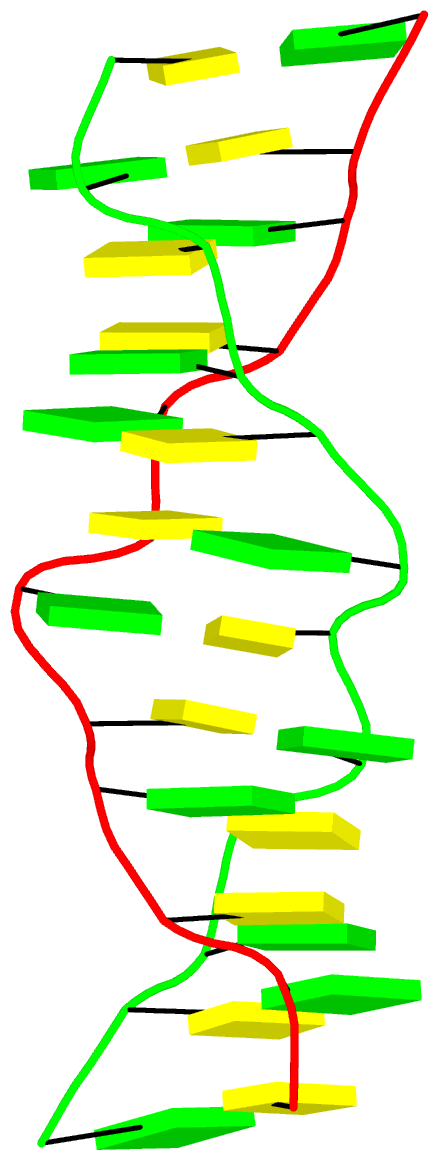

In January 29, 2025, I received the following email request from a long-time DSSR user:
... recently noted that 3DNA/DSSR automatically maps non-standard nucleotides to standard nucleotides. I wonder if you would be willing to share with us your most current version of mappings?
I responded to the user the same day, with detailed information about the mapping process in DSSR. The user was happy with my response, and that thread was quickly closed with a positive note.
On April 22, 2025, a related question, titled "Can x3dna-dssr correctly handle N1-methyl-pseudouridine?", was asked on the 3DNA Forum. In answering the question on the Forum, I referred to my email response to the previous user.
I now realize that writing a detailed blog post explaining the mapping process would be beneficial for DSSR users. It would also enable me to easily reference this blog post in future interactions with users.
3DNA/DSSR performs automatic mapping of modified nucleotides (including pseudouridine) to their standard counterparts. Over the years, the method has proven to work well in real-world applications. It is one of the defining features that make DSSR just work. For example, for the tRNA 1ehz, DSSR automatically identifies the following 14 modified nucleotides (of 11 unique types):
# x3dna-dssr -i=1ehz.pdb
List of 11 types of 14 modified nucleotides
nt count list
1 1MA-a 1 A.1MA58
2 2MG-g 1 A.2MG10
3 5MC-c 2 A.5MC40,A.5MC49
4 5MU-t 1 A.5MU54
5 7MG-g 1 A.7MG46
6 H2U-u 2 A.H2U16,A.H2U17
7 M2G-g 1 A.M2G26
8 OMC-c 1 A.OMC32
9 OMG-g 1 A.OMG34
10 PSU-P 2 A.PSU39,A.PSU55
11 YYG-g 1 A.YYG37
Users could run DSSR on a set of structures of interest, and collect the unique mappings for a complete list of modified nucleotides.
Moreover, DSSR has the --nt-mapping option that allows users to control the mapping process. The screenshot below is taken from the relevant part of the DSSR manual.
For example, DSSR automatically maps 5MU (5-methyluridine 5′-monophosphate) to t (i.e., modified thymine) because of the 5-methyl group. With the option --nt-mapping='5MU:u', DSSR would take 5MU as a modified uracil. This option allows for multiple mappings separated by comma. The mapping of 5MU to u or t is obviously arbitrary. DSSR is robust against the ambiguity in designating a modified nucleotide to its nearest canonical counterpart. For example, mapping 5MU to u or t has minimal influence on DSSR-derived base-pair parameters and other structural features.

Background information on the mapping
Over the years, I've refined the heuristics of the mapping process. In the early days with 3DNA, I kept an ever increasing list in file baselist.dat with hundreds of entries like: MIA a that maps MIA as a modified A, denoted as lowercase a. In recent releases of DSSR, I keep only the standard ones, with a total of 48 entries like ADE A, and DG5 G etc. If a residue is not a standard one, the following C function is called to do the mapping. DSSR performs filtering to decide if a residue is a nucleotide, and if so R (purine) or Y (pyrimidine).
static void derive_new_nt_std_name(long resi, struct_mol *pdb, char *info)
{
char str[BUF512];
double d1 = DMAX, d2 = DMAX;
long C1_prime, N1, C5;
struct_residue *r = &pdb->residues[resi];
if (r->type[RESIDUE_NT_UNKNOWN]) {
sprintf(r->std_name, "__%c", Gvars.abasic);
return;
}
if (is_R(resi, pdb)) { /* purine */
if (residue_has_atom(" O6 ", resi, pdb)) /* with ' O6 ' */
strcpy(r->std_name, "__g");
else if (!residue_has_atom(" N6 ", resi, pdb) && /* no ' N6 ' but ' N2 ' */
residue_has_atom(" N2 ", resi, pdb))
strcpy(r->std_name, "__g");
else
strcpy(r->std_name, "__a");
} else { /* a pyrimidine */
if (residue_has_atom(" N4 ", resi, pdb))
strcpy(r->std_name, "__c");
else if (residue_has_atom(" C7 ", resi, pdb))
strcpy(r->std_name, "__t");
else
strcpy(r->std_name, "__u");
C1_prime = find_atom_in_residue(" C1'", resi, pdb);
N1 = find_atom_in_residue(" N1 ", resi, pdb);
if (atoms_same_model_chain_altloc(C1_prime, N1, pdb))
d1 = dist_atoms(C1_prime, N1, pdb);
if (!dval_in_range(d1, 1.0, 2.0)) {
C5 = find_atom_in_residue(" C5 ", resi, pdb);
if (atoms_same_model_chain_altloc(C1_prime, C5, pdb))
d2 = dist_atoms(C1_prime, C5, pdb);
if (dval_in_range(d2, 1.0, 2.0))
strcpy(r->std_name, "__p");
}
}
if (!Gvars.standalone) {
sprintf(str, "\n\tmatched nucleotide '%s' to '%c' for %s\n"
"\tverify and add an entry in <baselist.dat>\n",
r->res_name, r->std_name[2], info);
logit(str);
}
}

Recently, I read carefully the two papers by Farag et al. on the ASC-G4 algorithm to calculate "advanced structural characteristics of G-quadruplexes" (2023), and the comprehensive analysis results of intramolecular G4 structures in the PDB (2024). By developing a convention to orient and number the four strands, ASC-G4 allows for unambiguous determination of the intramolecular G4 topology. It also has an in-depth discussion on assigning syn or anti glycosidic configuration of guanosines, and categorizes four different types of snapbacks.
I am glad to see that DSSR is cited in these two papers, as quoted below:
X3dna-DSSR (19) (http://x3dna.org) is a website that was created to calculate nucleic acid structural parameters, like the local base-pair parameters, local step base-pair parameters, torsion angles, etc, but not the special characteristics of G4. A subdomain dedicated to G4, DSSR-G4DB (Dissecting the Spatial Structure of RNA – G4 Data Base) (http://g4.x3dna.org) emanated from this website. It is a database that gathers and calculates some specific structural information about published G4s, like the topology, the rise, the helical twist, etc, but not the groove widths or the presence of snapbacks. -- Farag et al. (2023)
Indeed, DSSR-G4DB dose not classify snapbacks. I was aware of such non-canonical G4s when I first developed the G4 module in DSSR around 2017-2018, and the V-shaped loops was derived to reflect the peculiarity of snapbacks.
DSSR classifies groove widths as medium, wide, or narrow, based on the glycosidic angles of neighboring guanosines in a G-tetrad, following the G4 literature. Using PDB entry 2lod as an example, the relevant part of the DSSR output is shown below. The groove widths of the three G-tetrads in the G4-stem have the same pattern of groove=--wn, standing for medium, medium, wide, and narrow, respectively. Note that the medium groove is represented by a dash instead of m because --wn stands out more clearly than mmwn (similar idea applies to glycosidic bond, e.g., sss-).
1 glyco-bond=sss- sugar=---- groove=--wn Major-->WC N- nts=4 GGGG A.DG1,A.DG6,A.DG20,A.DG16
2 glyco-bond=---s sugar=---- groove=--wn WC-->Major N+ nts=4 GGGG A.DG2,A.DG7,A.DG21,A.DG15
3 glyco-bond=---s sugar=---- groove=--wn WC-->Major N+ nts=4 GGGG A.DG3,A.DG8,A.DG22,A.DG14
Since DSSR-G4DB is a database, the user cannot provide his own G4 structure, to obtain structural information. Hence the necessity of developing a website where the user uploads his G4 structure file to obtain all its important and specific structural characteristics (like the topology, the groove width, the tilt and twist angles, etc.). This can be very useful, not only for the analysis of published PDB structures but also for structures in refinement or obtained from MD simulations, to evaluate their quality. To our knowledge, there is no website dedicated to G4 to do such calculations in real-time. Therefore, we developed the algorithm ASC-G4 (advanced structural characteristics of G4) and deployed it as a user-friendly website at the following address: http://tiny.cc/ASC-G4. -- Farag et al. (2023)
Thanks to the NIH R24GM153869 grant support, the http://g4.x3dna.org website now allows users to upload their own atomic structures in PDB for mmCIF format for the identification, annotation, and visualization of G4s. See the example of uploading PDB coordinate file 2lod.pdb.
As background, I had long aspired to develop a dynamic website for on-demand G4 structural analysis but was unable to pursue this goal until recently. During the 4-year funding gap, I still managed to maintain the website g4.x3dna.org, which provides DSSR results for G4 structures in the PDB (a resource now known as the DSSR-G4DB database). To date, the only published work related to G4s is my 2020 paper on the integration of DSSR with PyMOL. Clearly, a dedicated method paper detailing the G4 module in DSSR and the g4.x3dna.org website has been long overdue.
As an initial step toward addressing this gap, I have recently revised the G4-related code in DSSR, fixed existing bugs, and added new features. The g4.x3dna.org website has undergone a complete overhaul, enabling users to upload their own structures for dynamic G4 analysis. Additionally, the DSSR-G4DB database is being actively updated on a weekly basis as new PDB entries are added.
Calculation of the twist and tilt angles. In G4, the helix twist is the rotation of a tetrad relative to its successive one. To measure the twist angle, the most spread method is that described by Lu and Olson (2003) (32) and Reshetnikov et al. (2010) (33). In this method, the angle is calculated from the dot product between two C1’–C1’ vectors from two successive tetrads, i and i + 1, the C1’ atoms of each vector belonging to two adjacent guanosines of a Hbp. The issue with this method is that it does not allow access to the sign of the angle, which defines the direction of the G4 helix, viz. right-handed or left-handed. -- Farag et al. (2023)
There is clearly a misunderstanding in the above text. 3DNA/DSSR can handle left-handed Z-DNA without any issues. DSSR also reports negative twist angles for left-handed G4s, as shown clearly for PDB entry 7d5e, for example.
3DNA/DSSR derives a complete of set of six base-pair parameters (including shear and opening), six step parameters (including twist and rise), and six helical parameters, using a rigorously defined and completely reversible algorithm (CEHS) and the standard base-reference frame. See section "3.2.3 Base pairs" in DSSR User Manual for more details. The DSSR output for G4s (as in DSSR-G4DB) reports only twist and rise, along with overlapped areas, simply because these are the most important parameters and easily interpretable.
The list of the resolved G4 structures was downloaded from the ONQUADRO website (https://onquadro.cs.put.poznan.pl/) (39) at about the end of October 2023. It consisted of 291 intramolecular structures (named unimolecular in the website) and 154 intermolecular G4s (96 bimolecular and 58 tetramolecular). Only the intramolecular structures were kept for this study. To this list, we added 55 missing intramolecular structures that were found on the website of DSSR-G4DB (http://g4.x3dna.org) (40). From the merged list, 345 structures were downloaded from the Protein Data Bank (PDB) (http://www.rscb.org/pdb/) (41) because one structure had no available coordinates in the PDB format (7ZJ5 (42)). -- Farag and Mouawad (2024)
DSSR adopts the frame of reference of Webba da Silva, designating the four strands and grooves of G4-stem as shown below using PDB entries 8ht7 (G1 in syc) and 5ua3 (G1 in anti) as example for the syn or anti glycosidic bond of the 5'-guanosine, respectively.
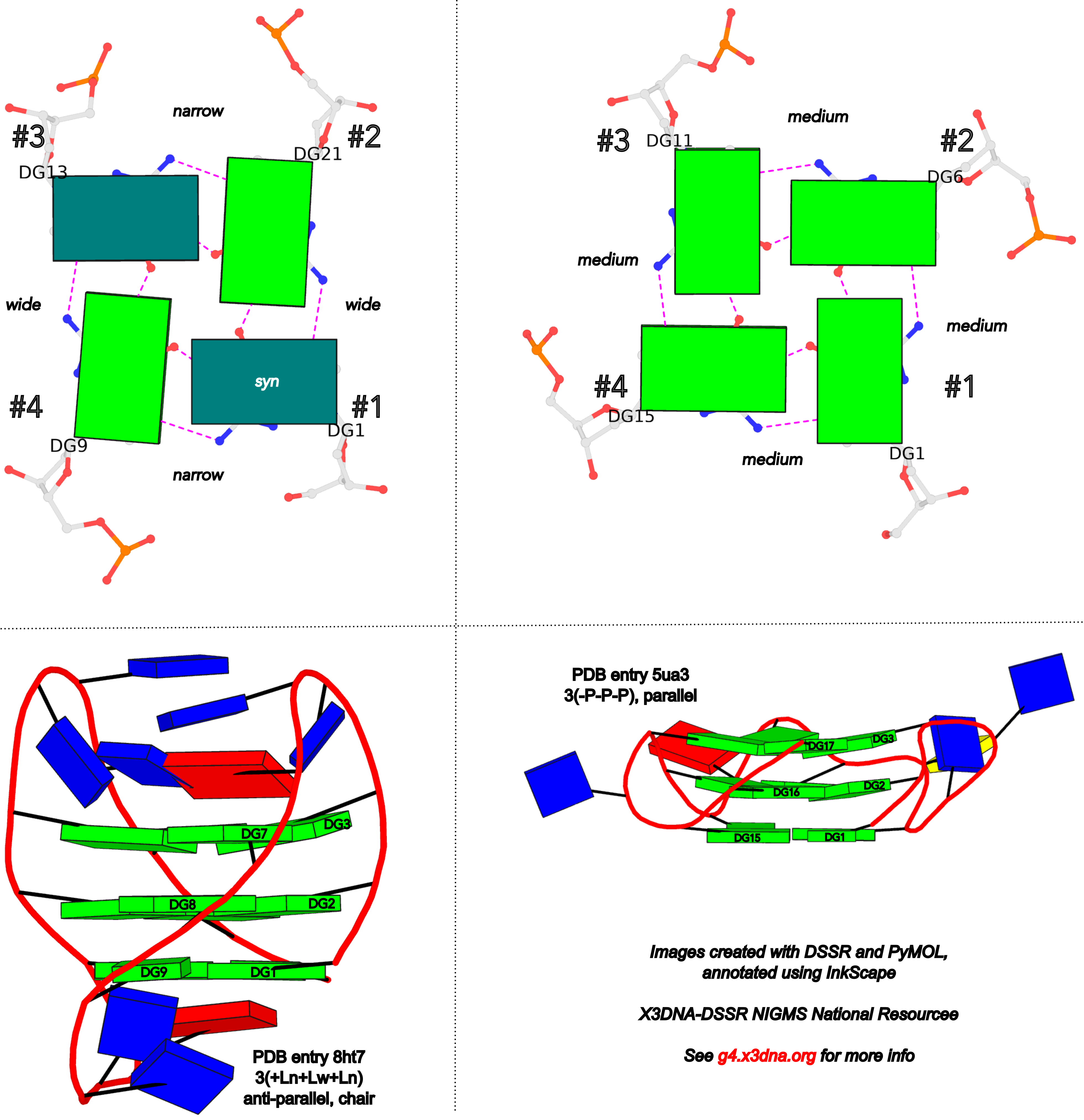
In DSSR, the first strand (#1) is always upward (U) from 5' to 3'-end, and the polarity of the other three strands is determined by its orientation relative to #1: U if parallel, or D if antiparallel. There are a total of 2x2x2=8 possible combinations of U and D for the three strands, which define parallel (U4: UUUU), antiparallel (U2D2: UDDU, UDUD, UUDD), or hybrid (UD3: UDDD; U3D: UDUU, UUDU, UUUD). For example, the PDB entry 2lod is characterized by DSSR as: "hybrid-(mixed), UUUD, U3D(3+1)", and PDB entry 8ht7 as: "anti-parallel, UDUD, chair(2+2)". This notation is topologically equivalent to the one adopted by ASC-G4 but with opposite orientation of the strands.
Overall, DSSR and ASC-G4 provide different perspectives on G4 structures. It is to the user to decide which one is more suitable for their needs.
References
Farag,M. et al. (2023) ASC-G4, an algorithm to calculate advanced structural characteristics of G-quadruplexes. Nucleic Acids Res., 51, 2087–2107.
Farag,M. and Mouawad,L. (2024) Comprehensive analysis of intramolecular G-quadruplex structures: furthering the understanding of their formalism. Nucleic Acids Res., gkae182.

In late September of 2018, I contacted Dr. Mateus Webba da Silva requesting a copy of his 2007 article, titled "Geometric formalism for DNA quadruplex folding". At that time, I had implemented a G4 module within DSSR for the automatic identification, comprehensive annotation, and schematic visualization of G-quadruplexes from 3D atomic coordinates. I noticed the 2007 paper, and was intrigued by the following sentences in the abstract:
A formalism is presented describing the interdependency of a set of structural descriptors as a geometric basis for folding of unimolecular quadruplex topologies. It represents a standard for interpretation of structural characteristics of quadruplexes, and is comprehensive in explicitly harmonizing the results of published literature with a unified language.
Mateus kindly sent me a copy of the 2007 article, and shortly afterwards he also shared with me the Dvorkin et al. (2018) paper on "Encoding canonical DNA quadruplex structure". I carefully read both papers, plus the Karsisiotis et al. (2013) tutorial paper. I was impressed by the elegance of the formalism: simple and systematic, so I immediately decided to add this feature to the G4 module of DSSR.
As the Chinese saying goes, "纸上得来终觉浅,绝知此事要躬行" ("What you learn from books is always shallow. You must practice it yourself to know it well." -- Google Translate). The implementation process was challenging because of subtleties in the formalism, but very rewarding. It is all about scientific understanding and software engineering. Only after a thorough understanding and attention to meticulous details can one create a robust and reliable software tool. On the other hand, once properly implemented, the DSSR G4 module can be applied consistently. Any discrepancies between DSSR output and literature merit further investigation. These discrepancies could either arise from bugs in DSSR (which I will promptly address upon identification) or, more likely, typos or errors in the reported results.
Webba da Silva (2007) systematically described the interdependency of glycosidic bond (syn or anti), strand polarity (parallel or anti-parallel), groove width (narrow, medium, or wide), and loop type (lateral, propeller, or diagonal) in unimolecular G-quadruplexes. Figures 1-3 and Scheme 1 of Webba da Silva (2007) are very informative, and easy to follow conceptually. The Karsisiotis et al. (2013) tutorial provided further details based on experimentally determined G-quadruplex structures from the PDB (e.g., Figure 3: the schematic for all possible combinations of glycosidic bond and the corresponding groove-width combinations in G-tetrad). Some key observations:
- Since glycosidic bond can be either syn or anti, there a total of
2x2x2x2 = 16 possible combinations in a G-tetrad.
- The disposition of glycosidic bond of guanosines in a G-tetrad leads to only eight possible groove-width combinations.
- Only tetrads with the same groove-width combinations may stack to form stable G-quadruplexes.
- Propeller loops invariably link medium grooves within a G-quadruplex stem.
- Lateral and diagonal loops bridge guanosines of different glycosidic bond.
- If a single-stranded quadruplex starts with a narrow groove, it can only be with a clockwise loop progression (i.e., +lateral).
- There are 26 permissible looping combinations within a canonical unimolecular G-quadruplex (G4-stem).
To unambiguously characterize a G4-stem, Webba da Silva (2007) defined a frame of reference where the 5’-G in a G4-stem is set as the origin, and the first strand is progressing towards the viewer. Regardless of the clockwise or anti-clockwise progression of the base sequence, the scheme designates one orientation for the syn and anti glycosidic bond by following G+G H-bonding alignments. Put another way, grooves and strands are strictly related to the reference (first) strand in an anti-clockwise manner, irrespective of the progression of the base sequence. The point is illustrated in the figure below, using PDB entries 8ht7 (G1 in syc) and 5ua3 (G1 in anti) as an example for the syn or anti glycosidic bond of the 5'-guanosine, respectively.

Based on previous work, the Dvorkin et al. (2018) paper proposed a systematic nomenclature for G4-stem. The single structural descriptor contains:
- The number of G-tetrads (i.e., the G-tract length).
- Loop types (lowercase l for lateral, p for propeller, and d for diagonal) and relative direction ("+" for clockwise and "-" for anti-clockwise progression, using the frame of reference described above).
- For lateral loops, the groove widths ("w" for wide, and “n” for narrow) are denoted in subscript.
So a complete descriptor could be 2(+lnd−p), as shown in Figure 1A of the Dvorkin et al. (2018) paper. Significantly, Figure 1B therein further gave structural descriptors for six experimentally determined G4-stems from the PDB. These examples, plus the ones in the supplementary materials, were used to validate my implementation of the systematic nomenclature in the G4 module of DSSR. My results agree with those in the Dvorkin et al. (2018) paper, except for two cases, which are discussed below.
- For PDB entry 2gku: 3(-p-ln-lw) (Dvorkin et al.) vs 3(-P-Lw-Ln) (DSSR), with swapped n (narrow) and w (wide) groove widths for both lateral loops.
- For PDB entry 2lod: 3(-pd+ln) (Dvorkin et al.) vs 3(-PD+Lw) (DSSR), with swapped n (narrow) and w (wide) groove width for the lateral loop.
Note that in DSSR, I am using uppercase L/P/D for lateral/propeller/diagonal loop types, and lowercase n/w for narrow/wide groove widths, respectively. Doing so distinguishes between the different loop types and groove widths in pure text format.
After careful examination of these discrepancies, I still couldn’t find any errors in my implementation. So I contacted Mateus for verification (in early October 2018). Thankfully, he quickly responded and acknowledged the mistakes for PDB entry 2gku in Dvorkin et al. (2018), saying "There can not be a –Ln after the –p." Clearly, the wrong descriptor for PDB entry 2gku in Dvorkin et al. (2018) was due to a typographical error. This example illustrates the power of a robust software tool like DSSR.
References
Dvorkin,S.A. et al. (2018) Encoding canonical DNA quadruplex structure. Sci. Adv., 4, eaat3007.
Karsisiotis,A.I. et al. (2013) DNA quadruplex folding formalism – A tutorial on quadruplex topologies. Methods, 64, 28–35.
Webba da Silva,M. (2007) Geometric formalism for DNA quadruplex folding. Chemistry A European J, 13, 9738–9745.

Over the past few years, the Szachniuk group has made several significant contributions to the field of structural bioinformatics of G-quadruplexes. The following five publications are particularly noteworthy, and I am glad to see that 3DNA/DSSR have been cited in all of them.
1. Zok et al. (2020) -- ElTetrado: a tool for identification and classification of tetrads and quadruplexes
The BMC Bioinformatics paper introduced the ElTetrado software tool for identifying and classifying G-tetrads in unimolecular G-quadruplex structures into ONZ taxonomy. Here DSSR is employed to identify base-pairs and base-stacking interactions.
ElTetrado processes PDB and mmCIF files to identify quadruplexes and their component tetrads in nucleic acid structures (Fig. 2). It applies DSSR [24] to collect the preliminary information about base pairs and stacking.
We recommend that, apart from ElTetrado, the users should download the DSSR binary [24] and place it in the same local directory. DSSR is utilized for the preliminary analysis of base pairs in the input 3D structure. Its local installation allows the users to control DSSR execution. For example, one can decide to pass --symmetry parameter to x3dna-dssr binary when dealing with X-ray structures, which is necessary for some quadruplexes.
As documented in the DSSR Manual, by default, DSSR reads in the first model of an NMR ensemble. A biological unit of X-ray crystal structures in the PDB may contain symmetry-related components formatted as a MODEL/ENDMDL delimited, NMR-like ensemble. In such cases, the --symmetry (or --symm) option is required for DSSR to process the entire biological unit.
For example, x3dna-dssr -i=4ms9.pdb1 --symm leads to the identification of 10 Watson-Crick base pairs in the biological unit of PDB entry 4ms9 (uploaded). The --symm option is now enabled for user-uploaded PDB files on the skmatic.x3dna.org website. Without the --symm option, DSSR would not find any Watson-Crick base pairs in 4ms9.pdb1 since MODEL#1 is single stranded.
Noticing the confusion users may have in using the --symm option, I have revised DSSR to check for overlapped residues. When all models in an NMR ensemble are taken as a whole with the --symm option, there will be overlapped residues. In such cases, DSSR will report a diagnostic message and proceed with the first model only. The final result is as if --symm has not been specified. Put another way, specifying --symm for an NMR ensemble does no harm to the analysis. For example, analyzing PDB entry 8xeq with the --symm option would have the following message and only the first model would be processed.
x3dna-dssr -i=8xeq.pdb --symm -o=8xeq.out
[i] You specified --symm, but the input file is an (NMR) ensemble
*** in the following, only the FIRST model will be processed ***
Alternatively, if the users do not want to have a local version of DSSR binary, they can obtain the DSSR output in JSON format from any place and use them as input data for ElTetrado (with --dssr-json parameter).
ElTetrado is started from the command line. The users enter the program name and either --pdb followed by an input file name (the file should be in PDB or mmCIF format), or --dssr-json followed by a path to JSON file generated by DSSR, or both switches at once.
The JSON output from DSSR can be obtained directly from the skmatic.x3dna.org website for pre-processed PDB entries or user-supplied coordinate files. For examples, for PDB entry 1ehz, the URL is http://skmatic.x3dna.org/pdb/1ehz/1ehz.json. Alternatively, users can use the web API to get the JSON file, as shown below:
# Pre-processed PDB entry:
curl http://skmatic.x3dna.org/api/pdb/1ehz/json
# With user-supplied PDB file
curl http://skmatic.x3dna.org/api -F 'model=@1ehz.pdb' -F 'type=json'
curl http://skmatic.x3dna.org/api -F 'url=https://files.rcsb.org/download/1ehz.pdb.gz' -F 'type=json'
2. Popenda et al. (2020) -- Topology-based classification of tetrads and quadruplex structures
This paper presents the ONZ scheme to clarify tetrads in unimolecular structures (see Figure 2 therein). Note that DSSR, with option --G4=ONZ, classifies G-tetrads into ONZ taxonomy. For example, the PDB entry 2gku has one G-tetrad (G3, G9, G17, G21) in the O- category, and two G-tetrads in O+.
Structures from both sets were analyzed using self-implemented programs along with DSSR software from the 3DNA suite (Lu et al., 2015). From DSSR, we acquired the information about base pairs and stacking.
3. Zurkowski et al. (2022) -- DrawTetrado to create layer diagrams of G4 structures
DrawTetrado generates static layer diagrams that represent structural data in a pseudo-3D perspective. The layer diagram is very informative and visually pleasing, and it complements the cartoon block schematics generated by the DSSR-PyMOL integration and the detailed DSSR characterization of G-quadruplexes.
So far, the only visual model designed for the 3D structure of quadruplexes is cartoon block schematics (Fig. 1C). These models are generated by DSSR-PyMOL integration and presented as static images of the structure viewed from six perspectives (Lu, 2020).
4. Adamczyk et al. (2023) -- WebTetrado: a webserver to explore quadruplexes in nucleic acid 3D structures
The topologies underlying the classification of quadruplexes and other parameters of their structures can be analyzed using a few computational tools. DSSR (7) was the first to target the detection of G-quadruplexes in 3D structure data saved in PDB and PDBx/mmCIF files and to describe their features. It runs systematically on all entries in the Protein Data Bank and collects motifs found in the DSSR-G4DB database. ElTetrado (8) can identify and analyze G4s and other kinds of tetrads and quadruplexes, classify them, and compute their parameters. It is the core of the computation pipeline running within the ONQUADRO database system (9). The most recent tool for processing atom coordinates in the search for quadruplexes is ASC-G4 (10). It calculates more features than DSSR and ElTetrado, but is limited to unimolecular quadruplexes and supports only the PDB format.
The example illustration in Figure 3 of the paper is on PDB entry 6h1k, the major G-quadruplex form of HIV-1 LTR (long terminal repeat). The layer diagram shown below, re-generated using the WebTetrado website, helps visualize the detailed characterization of DSSR very nicely.
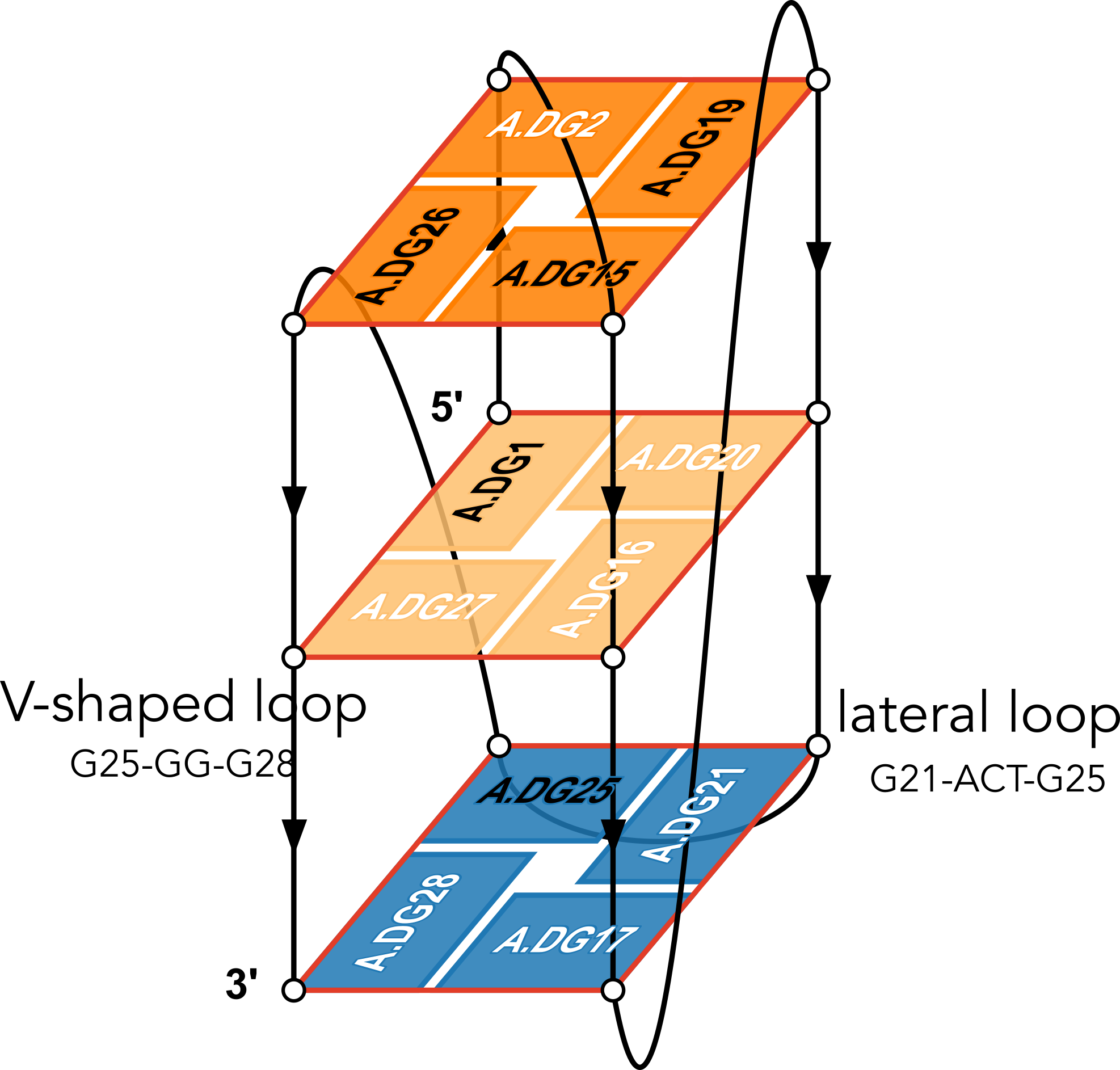 "In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem." For PDB entry 6h1k, DSSR identifies a G-helix with three G-tetrads, ordered properly. Specifically, strand#1 consists of (G2, G1, and G25), even G1 and G25 are not covalently connected. On the other hand, "In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands." Thus, the G4-stem is composed of only two G-tetrads, as detailed below.
"In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem." For PDB entry 6h1k, DSSR identifies a G-helix with three G-tetrads, ordered properly. Specifically, strand#1 consists of (G2, G1, and G25), even G1 and G25 are not covalently connected. On the other hand, "In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity. Bulges are also allowed along each of the four strands." Thus, the G4-stem is composed of only two G-tetrads, as detailed below.
Stem#1, 2 G-tetrads, 3 loops, INTRA-molecular, UDDD, hybrid-(mixed), 2(D+PX), UD3(1+3)
1 glyco-bond=s--- sugar=---- groove=w--n Major-->WC Z- nts=4 GGGG A.DG1,A.DG20,A.DG16,A.DG27
2 glyco-bond=-sss sugar=.-.3 groove=w--n WC-->Major Z+ nts=4 GGGG A.DG2,A.DG19,A.DG15,A.DG26
step#1 mm(<>,outward) area=12.76 rise=3.47 twist=18.2
strand#1 U DNA glyco-bond=s- sugar=-. nts=2 GG A.DG1,A.DG2
strand#2 D DNA glyco-bond=-s sugar=-- nts=2 GG A.DG20,A.DG19
strand#3 D DNA glyco-bond=-s sugar=-. nts=2 GG A.DG16,A.DG15
strand#4 D DNA glyco-bond=-s sugar=-3 nts=2 GG A.DG27,A.DG26
loop#1 type=diagonal strands=[#1,#3] nts=12 GAGGCGTGGCCT A.DG3,A.DA4,A.DG5,A.DG6,A.DC7,A.DG8,A.DT9,A.DG10,A.DG11,A.DC12,A.DC13,A.DT14
loop#2 type=propeller strands=[#3,#2] nts=2 GC A.DG17,A.DC18
loop#3 type=diag-prop strands=[#2,#4] nts=5 GACTG A.DG21,A.DA22,A.DC23,A.DT24,A.DG25
List of 2 non-stem G4-loops (including the two closing Gs)
1 type=lateral helix=#1 nts=5 GACTG A.DG21,A.DA22,A.DC23,A.DT24,A.DG25
2 type=V-shaped helix=#1 nts=4 GGGG A.DG25,A.DG26,A.DG27,A.DG28
DSSR correctly identifies the 12-nt diagonal loop, containing a canonical duplex stem and a hairpin loop. Notably, the G-tetrad (25-21-17-28) does not belong to the G4-stem because of the broken backbone connectivity between G1 and G25. Instead, the Gs in the G-tetrad (25-21-17-28) become part of the following two loops, which are certainly unconventional yet follow naturally the DSSR definition of G4-stem.
- The propeller loop, which now includes G17 (part of the G-tetrad), in addition to C18.
- The unusual diag-prop (diagonal-propeller ) loop, which consists of G21, A22, C23, T24, and G25.
Moreover, DSSR also reports two loops that are not defined by the G4-stem: the V-shaped loop (G25-G26-G27-G28) and the lateral loop (G21-A22-C23-T24-G25). See the notes in the above layer diagram. V-shaped loop occurs when the 5’-endmost G-tetrad lies in the middle of the G-quartets stack as in the non-canonical G4 structures with snapbacks.
The G4-helix and G4-stem definitions parallel those for duplex helix and stem in DSSR. The characterization of loops follows naturally once G4-stem or duplex stem are identified. The unusual propeller and diagonal-propeller loops noted above are due to non-canonical structures, which also lead to the listing of non-stem G4-loops.
I may consider to add special handling of snapbacks (or other worthwhile classes of non-canonical G4 structures) so that the reported loops follow whatever consensus the community agrees upon in the future. Nevertheless, I would like to emphasize that the consistent definitions of G4-stem and loops in DSSR help pinpoint extraordinary features to draw users' attention to non-canonical G4 structures. The layer diagram from DrawTetrado and WebTetrado are very handy in illuminating the basic concept and technical details, as shown here for PDB entry 6h1k.
5. Zok et al. (2022) -- ONQUADRO: a database of experimentally determined quadruplex structures
The computational engine is composed of scripts utilising in-house and third-party procedures, responsible for data collection, quadruplex identification, computation of structure parameters, secondary structure annotation, visualisation of the secondary and tertiary structure models, database queries, generation of statistics, and newsletter preparation. DSSR (--pair-only mode) (36) and ElTetrado (39) functionalities are applied to identify quadruplexes, tetrads, and G4-helices in nucleic acid structures.
References
Adamczyk, B., Zurkowski, M., Szachniuk, M., & Zok, T. (2023). WebTetrado: a webserver to explore quadruplexes in nucleic acid 3D structures. Nucleic Acids Research, 51(W1), W607–W612. https://doi.org/10.1093/nar/gkad346
Popenda, M., Miskiewicz, J., Sarzynska, J., Zok, T., & Szachniuk, M. (2020). Topology-based classification of tetrads and quadruplex structures. Bioinformatics, 36(4), 1129–1134. https://doi.org/10.1093/bioinformatics/btz738
Zok, T., Kraszewska, N., Miskiewicz, J., Pielacinska, P., Zurkowski, M., & Szachniuk, M. (2022). ONQUADRO: a database of experimentally determined quadruplex structures. Nucleic Acids Research, 50(D1), D253–D258. https://doi.org/10.1093/nar/gkab1118
Zok, T., Popenda, M., & Szachniuk, M. (2020). ElTetrado: a tool for identification and classification of tetrads and quadruplexes. BMC Bioinformatics, 21(1), 40. https://doi.org/10.1186/s12859-020-3385-1
Zurkowski, M., Zok, T., & Szachniuk, M. (2022). DrawTetrado to create layer diagrams of G4 structures. Bioinformatics, 38(15), 3835–3836. https://doi.org/10.1093/bioinformatics/btac394

I recently came across the Direk & Doluca (2024) paper on CIIS‐GQ: Computational Identification and Illustrative Standard for representation of unimolecular G‐Quadruplex secondary structures. Since DSSR is mentioned extensively in this work, with a section comparing CIIS-GQ and DSSR in supplementary materials, it is worthwhile to explore the issues raised in the paper. Overall, following literature allows me to clarify misconceptions and fix bugs that further improve DSSR.
The data which contain the G-quadruplexes were identified by DSSR-G4DB website [12, 13, 16]. All of the PDB (protein data bank) ids of DNA and RNA structures are extracted from the aforementioned website and the pdb files which contain the three dimensional data of the corresponding structures were downloaded from the protein data bank [3–5]. [under section "Materials and Methods": "Data"]
The DNA and RNA structures listed in 3DNA website were identified and downloaded from Protein Data Bank. Only unimolecular structures were used for the rest of the study (Supplementary Fig. 2). [under section "Results"]
Additionally, DSSR requires licensing to get annotation results for G-quadruplex structures. Fortunately, the annotation results for a number of G-quadruplexes were already published at DSSR-G4DB (46) and we were able to compare. [under section "Comparison with DSSR" in supplemental materials]
I am glad the DSSR-G4DB website served as a starting point for this study. The G4.x3dna.org website, where DSSR-G4DB is hosted, has always been available to the public. With the NIH R24GM153869 grant support, the standalone DSSR software is free for academic use and can be obtained from the Columbia Technology Ventures (CTV) website.
All obtained results for each pdb file were compared with DSSR. Out of which 35 DNA and 13 RNA structures were analyzed differently (Supplementary Table 2). Significant differences were detected for a number of structures between CIIS-GQ and DSSR analysis. For example, in 1k8p, 3ibk, 6ip7 and 5ccw structures, DSSR fails to identify some loops in some structures.
Most common issue that we have observed with DSSR is that it places loops in wrong places in some structures. For example, In 2a5p structure, the first loop is identified as reversal by both tools but DSSR also assigns the G6 to this loop which already participates in a tetrad. Such misplacement of tetrad-forming guanines in a loop is also seen in other structures such as 2a5r, 2kpr, 2m53, 2m92, etc (Supplementary Table 2).
The G4 module in DSSR was first developed around 2017-2018 and the work was mentioned briefly in the Lu (2020) paper on DSSR-PyMOL integration. However, due to the funding gap, the development of the G4 module was put on hold. I have never got a chance to write a paper documenting the detailed algorithms for the identification, annotation, and visualization of G-quadruplexes. I recently revamped the G4.x3dna.org website from inside out, and reprocessed all PDB structures to compile the DSSR-G4DB database. Along the way, the G4 module has been updated and improved. Now I'm actively working on a manuscript on the G4 module in DSSR and the associate website.
DSSR has clear definitions of G4-helix and G4-stem, and the corresponding loops. Specifically, for PDB entry 2a5p, DSSR reports the following:
## List of 1 G4-helix
In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity,
and may contain more than one G4-stem.
##### Helix#1, 3 G-tetrads, INTRA-molecular, with 1 stem
1 glyco-bond=---- sugar=---- groove=---- WC-->Major O+ nts=4 GGGG A.DG4,A.DG8,A.DG13,A.DG17
2 glyco-bond=---- sugar=.--- groove=---- WC-->Major O+ nts=4 GGGG A.DG5,A.DG9,A.DG14,A.DG18
3 glyco-bond=-s-- sugar=-3-- groove=wn-- WC-->Major Z- nts=4 GGGG A.DG6,A.DG24,A.DG15,A.DG19
step#1 pm(>>,forward) area=9.64 rise=3.19 twist=32.7
step#2 pm(>>,forward) area=12.93 rise=3.29 twist=29.4
strand#1 DNA glyco-bond=--- sugar=-.- nts=3 GGG A.DG4,A.DG5,A.DG6
strand#2 DNA glyco-bond=--s sugar=--3 nts=3 GGG A.DG8,A.DG9,A.DG24
strand#3 DNA glyco-bond=--- sugar=--- nts=3 GGG A.DG13,A.DG14,A.DG15
strand#4 DNA glyco-bond=--- sugar=--- nts=3 GGG A.DG17,A.DG18,A.DG19
Notice the differences in grooves between the first two G-tetrads vs the 3rd one, and the breaking backbone for strand#2 between G9 and G24.
## List of 1 G4-stem
In DSSR, a G4-stem is defined as a G4-helix with backbone connectivity.
Bulges are also allowed along each of the four strands.
##### Stem#1, 2 G-tetrads, 3 loops, INTRA-molecular, UUUU, parallel, 2(-P-P-P), parallel(4+0)
1 glyco-bond=---- sugar=---- groove=---- WC-->Major O+ nts=4 GGGG A.DG4,A.DG8,A.DG13,A.DG17
2 glyco-bond=---- sugar=.--- groove=---- WC-->Major O+ nts=4 GGGG A.DG5,A.DG9,A.DG14,A.DG18
step#1 pm(>>,forward) area=9.64 rise=3.19 twist=32.7
strand#1 U DNA glyco-bond=-- sugar=-. nts=2 GG A.DG4,A.DG5
strand#2 U DNA glyco-bond=-- sugar=-- nts=2 GG A.DG8,A.DG9
strand#3 U DNA glyco-bond=-- sugar=-- nts=2 GG A.DG13,A.DG14
strand#4 U DNA glyco-bond=-- sugar=-- nts=2 GG A.DG17,A.DG18
loop#1 type=propeller strands=[#1,#2] nts=2 GT A.DG6,A.DT7
loop#2 type=propeller strands=[#2,#3] nts=3 gGA A.DI10,A.DG11,A.DA12
loop#3 type=propeller strands=[#3,#4] nts=2 GT A.DG15,A.DT16
Thus the G4-stem consists of two G-tetrads only, and G6 which is part of the 3rd G-tetrad becomes part of a propeller loop. Similar arrangement applies to the other cases.
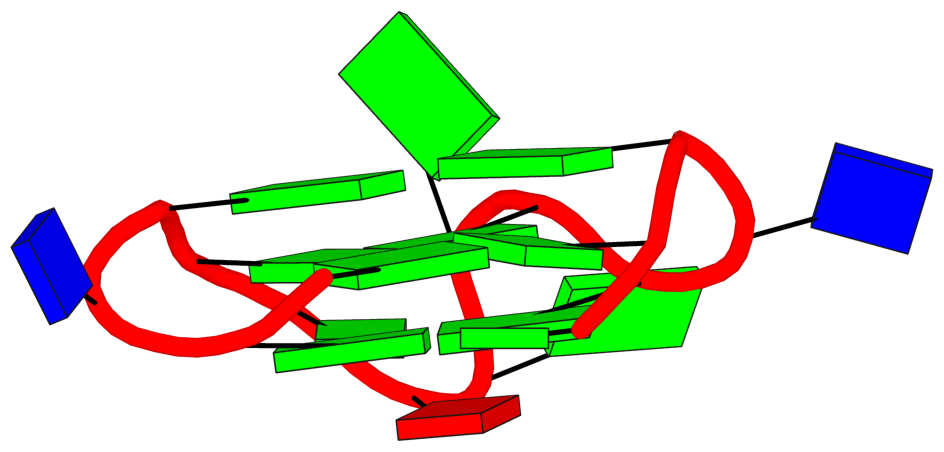 DSSR also reports the following loop:
DSSR also reports the following loop:
## List of 1 non-stem G4-loop (including the two closing Gs)
1 type=diagonal helix=#1 nts=6 GGAAGG A.DG19,A.DG20,A.DA21,A.DA22,A.DG23,A.DG24
In my understanding, the definition and nomenclature of loops in G4 structures are not yet standardized. I am monitoring the development in this field and will update DSSR as needed in due course.
There may also be different types of loops identified by these tools. For example, in 1oz8, which is depicted by CIISGQ as two separate G4s, DSSR fails to identify the G-tetrad, [2, 5, 8, 11], that lies on the outside of the structure. This results in identification of loops formed within this tetrad and its stacking neighbor different to CIIS-GQ. While CIISGQ identifies these loops as reversal, just like the other loops in the structure, DSSR identifies them as non-stem lateral loops. This causes complete misinterpretation of the size and the type of loops in the structure.
The revised DSSR output for PDB entry 1oz8 has the G-tetrad A.DG2,A.DG5,A.DG8,A.DG11 manually added as part of the input, and now all three propeller loops are correctly identified. By default, G11 does not form proper G+G pairs (of LW type cWH or cHW, and Saenger type VI) with G2 and G8. The distortion of the G-tetrad is obvious in the block representation of the structure.
In 4u5m, similar to 1oz8, the structure may be interpreted as two separate G4s connected through a single link (T13,T14). In this case, DSSR identifies only two loops in one of the G4s and labels them as non-stem V-shaped loops. This also differs from CIIS-GQ where CIIS-GQ interprets all loops in both G4 as reversal. Structures containing multiple G4s, such as 1oz8, 4u5m and 6kvb, are often identified with different loop types by DSSR, while CIIS-GQ can recognise the loops correctly and simplifies the comprehension of the structure.
For PDB entry 4u5m, the same arguments above regarding the G4-stem and loops for 2a5p apply.
1 glyco-bond=s--- sugar=---- groove=w--n Major-->WC O+ nts=4 GGGG A.DG2,A.DG11,A.DG8,A.DG5
2 glyco-bond=---- sugar=---- groove=---- Major-->WC O+ nts=4 GGGG A.DG3,A.DG12,A.DG9,A.DG6
3 glyco-bond=---- sugar=---- groove=---- WC-->Major O+ nts=4 GGGG A.DG24,A.DG15,A.DG18,A.DG21
4 glyco-bond=---- sugar=---- groove=---- WC-->Major O+ nts=4 GGGG A.DG23,A.DG26,A.DG17,A.DG20
As shown, the backbone between G15 and G26 is broken. Moreover, here the assignment of Gs along the strand may need to be manually adjusted.
As shown in Table 1, by relaxing angle and distance parameters, we were able to identify more tetrads (6T2G, 1OZ8) than DSSR, which detects them as multiplets instead.
The current DSSR results for PDB entry 6t2g and 1oz8 are all as expected. Moreover, DSSR can handle PDB entry 6t2g automatically, while for PDB entry 1oz8 user needs to manually edit the input to include the G-tetrad with G11. By allowing users to specify tetrads, DSSR offers precise control and great flexibility, e.g., to include the G-C-G-C tetrads in PDB entry 1a6h.
DSSR has a detailed explanation of strands, tetrads and loops. However, the comprehensive output of DSSR is often hard to understand and grasp the details of the structure. [in supplemental materials]
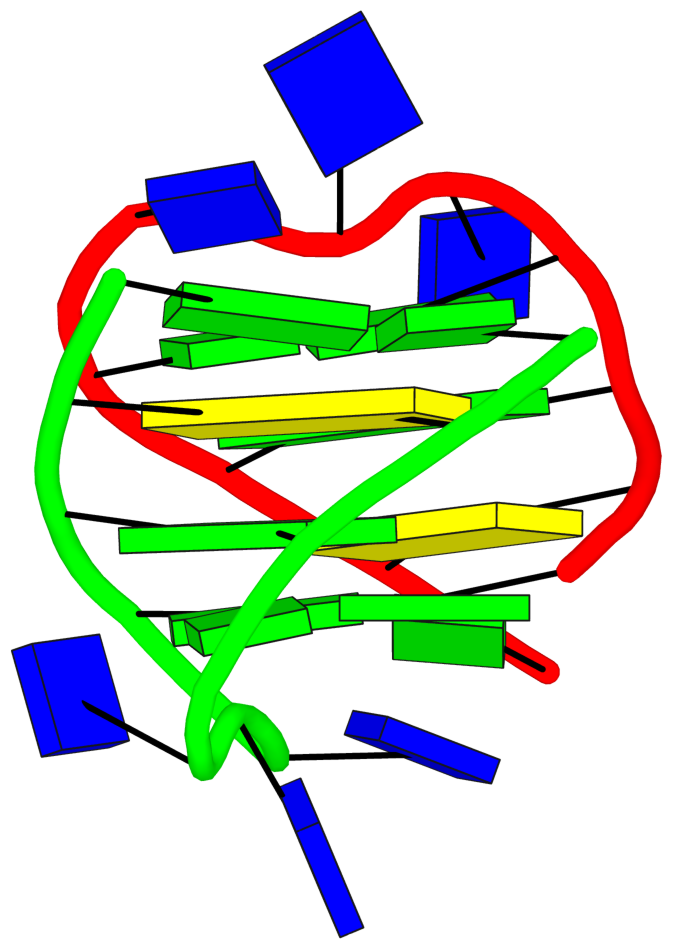
The detailed explanations are provided to help users understand the DSSR output. They are most insightful in combination with the schematic block diagrams. For examples, for PDB entry 1a6h, the middle G-C-G-C tetrads are crystal clear with the long green and yellow rectangular blocks, specially along with the detailed annotations of the tetrads, as shown below.
1 glyco-bond=s-s- sugar=---. groove=wnwn Major-->WC -- nts=4 GGGG A.DG1,A.DG11,B.DG8,B.DG4
2 glyco-bond=---- sugar=-.-- groove=---- -- -- nts=4 CGCG A.DC2,A.DG10,B.DC9,B.DG3
3 glyco-bond=---- sugar=--.- groove=---- -- -- nts=4 GCGC A.DG3,A.DC9,B.DG10,B.DC2
4 glyco-bond=-s-s sugar=---- groove=wnwn WC-->Major -- nts=4 GGGG A.DG4,A.DG8,B.DG11,B.DG1
Another advantage of CIIS-GQ is that it requires only two thresholds, the thresholds of distance and angle parameters that can be modified to detect loosely connected tetrads. Due to this advantage, the identification of the tetrads were possible in at least two structures. In case of 1OZ8, DSSR found three tetrads (G1-G4-G7-G10, G13-G16-G19-G22 and G14-G17-G20-G23) as shown at the result page2 (47) while CIIS-GQ has found one more tetrad which is G2-G5-G8-G11. In comparison DSSR highlighted G5-G8-G11 as a multiplet, omitting the G2. Based on this difference, loop classification differs with CIIS-GQ. DSSR has identified 3 stem reversal loops and 3 non-stem lateral loops while we have identified 7 reversal loops. Stem loop is defined as any loop that also forms a duplex within itself.
DSSR now has PDB entry 1oz8 properly characterized, by manually adding the G-tetrad involving G11, as detailed above.
A similar difference exists in 6T2G. DSSR could find 2 tetrads in this structure (G2-G6-G11-G26 and G4-G9-G13-G28) as shown at the result page3 (47) while CIIS-GQ found one more tetrad, G3-G7-G12-G27. DSSR is able to show these four guanines as a multiplet in the list of multiplets section, however does not present it as a tetrad like the other two tetrads. As a result, CIIS-GQ loop types and placements are also different. DSSR has found six lateral loops while CIIS-GQ has found three reversal loops.
DSSR can now handle PDB entry 6t2g automatically. Previous versions of DSSR missed the G-tetrad (G3+G7+G12+G27) because of the G12+G27 pair: it fails the criteria to be classified as the pair of LW type cWH or cHW and Saenger type VI. Thus G3+G7+G12+G27 do not qualify as a G-tetrad, but they still form a multiplet with four guanines.
References
Direk, T., & Doluca, O. (2024). Computational Identification and Illustrative Standard for Representation of Unimolecular G-Quadruplex Secondary Structures (CIIS-GQ). Journal of Computer-Aided Molecular Design, 38(1), 35. https://doi.org/10.1007/s10822-024-00573-1
Lu, X.-J. (2020). DSSR-enabled innovative schematics of 3D nucleic acid structures with PyMOL. Nucleic Acids Research, gkaa426. https://doi.org/10.1093/nar/gkaa426

The legacy PDB format has a field called “altLoc” (alternate location indicator) for "ATOM/HETATM" records in the "Coordinate Section". The corresponding documentation is excerpted below:
COLUMNS DATA TYPE FIELD DEFINITION
-----------------------------------------------------------------------
17 Character altLoc Alternate location indicator.
-
AltLoc is the place holder to indicate alternate conformation. The alternate conformation can be in the entire polymer chain, or several residues or partial residue (several atoms within one residue). If an atom is provided in more than one position, then a non-blank alternate location indicator must be used for each of the atomic positions. Within a residue, all atoms that are associated with each other in a given conformation are assigned the same alternate position indicator. There are two ways of representing alternate conformation- either at atom level or at residue level (see examples).
- For atoms that are in alternate sites indicated by the alternate site indicator, sorting of atoms in the ATOM/HETATM list uses the following general rules:
- In the simple case that involves a few atoms or a few residues with alternate sites, the coordinates occur one after the other in the entry.
- In the case of a large heterogen groups which are disordered, the atoms for each conformer are listed together.
In mmCIF format, AltLoc is under the atom_site category, with attribute name label_alt_id: i.e., labelled as _atom_site.label_alt_id. It is a required data item and appears in 43% of entries in the PDB.
In 3DNA and DSSR, AltLoc has a default value of "A1 ": an atom is taken into consideration if its AltLoc field (a single character) is space, A, or 1, otherwise it is ignored. Note that for mmCIF format, AltLoc field with dot (.) or question mark (?) is taken as space. Customized AltLoc values can be set via the --altloc option in DSSR.
Here is an example. PDB entry 7o1h is a 31-mer synthetic construct, with a hybrid-2R quadruplex-duplex of 3(-P-P-Lw) topology and three syn guanosines. It contains two modified residues designated BGM (BGM26 and BGM28), 8-bromo-2'-deoxyguanosine-5'-monophosphate, with AltLoc set to B. By default, DSSR detects only one G-tetrad, consisting of DG5,DG9,DG13,DG27, ignoring the two G-tetrads with BGM26 and BGM28. With the --altloc=B option (space is always included), all three G-tetrads are detected and the G-quadruplex (a G4-stem) is then automatically annotated as 3(-P-P-Lw):
# x3dna-dssr -i=7o1h-assembly1.cif --altloc=B
List of 2 types of 3 modified nucleotides
nt count list
1 BGM-g 2 A.BGM26,A.BGM28
2 THM-t 1 A.THM1
List of 1 G4-stem
Note: a G4-stem is defined as a G4-helix with backbone connectivity.
Bulges are also allowed along each of the four strands.
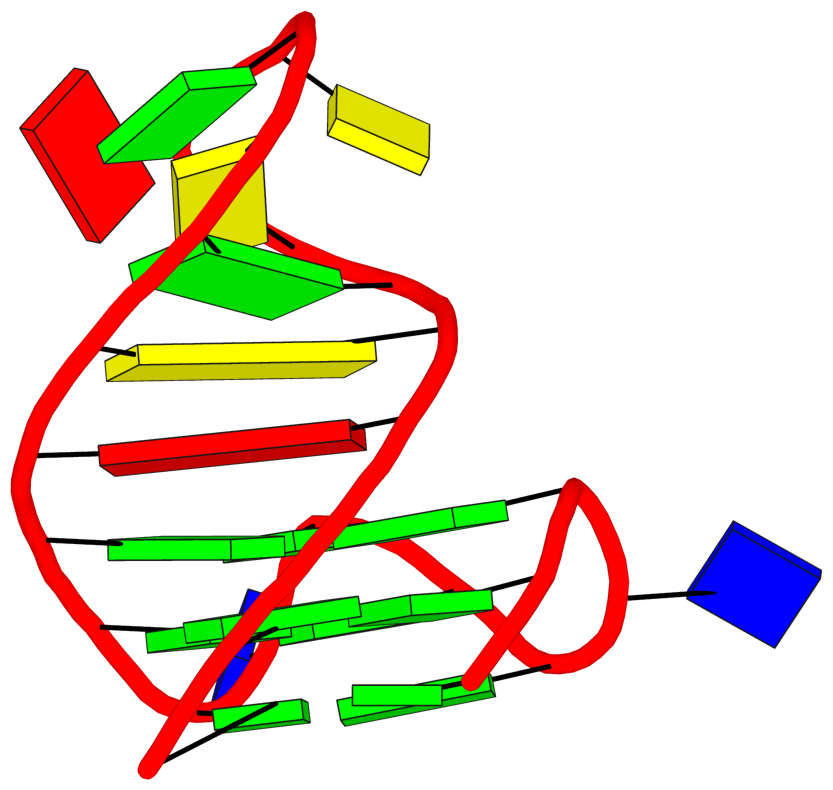
stem#1[#1] layers=3 INTRA-molecular loops=3 descriptor=3(-P-P-Lw) note=hybrid-2R(3+1) UUUD hybrid-(mixed)
1 glyco-bond=---s sugar=---. groove=--wn WC-->Major nts=4 GGGg A.DG4,A.DG8,A.DG12,A.BGM28
2 glyco-bond=---s sugar=--.- groove=--wn WC-->Major nts=4 GGGG A.DG5,A.DG9,A.DG13,A.DG27
3 glyco-bond=---s sugar=---- groove=--wn WC-->Major nts=4 GGGg A.DG6,A.DG10,A.DG14,A.BGM26
step#1 pm(>>,forward) area=13.57 rise=3.39 twist=26.7
step#2 pm(>>,forward) area=12.00 rise=3.44 twist=28.4
strand#1 U DNA glyco-bond=--- sugar=--- nts=3 GGG A.DG4,A.DG5,A.DG6
strand#2 U DNA glyco-bond=--- sugar=--- nts=3 GGG A.DG8,A.DG9,A.DG10
strand#3 U DNA glyco-bond=--- sugar=-.- nts=3 GGG A.DG12,A.DG13,A.DG14
strand#4 D DNA glyco-bond=sss sugar=.-- nts=3 gGg A.BGM28,A.DG27,A.BGM26
loop#1 type=propeller strands=[#1,#2] nts=1 T A.DT7
loop#2 type=propeller strands=[#2,#3] nts=1 T A.DT11
loop#3 type=lateral strands=[#3,#4] nts=11 ACGCGCAGCGT A.DA15,A.DC16,A.DG17,A.DC18,A.DG19,A.DC20,A.DA21,A.DG22,A.DC23,A.DG24,A.DT25
See G4.x3dna.org for DSSR-enabled annotation and visualization of this G4 structure. Here is the G4-stem in the frame of reference of 5' DG4 (bottom right), following the convention of Dvorkin et al. (2018). It is orientated automatically based on the standard base-reference frame (Olson et al. (2001)) of DG4.
References:
- Dvorkin, Scarlett A., Andreas I. Karsisiotis, and Mateus Webba Da Silva. 2018. “Encoding Canonical DNA Quadruplex Structure.” Science Advances 4 (8): eaat3007. https://doi.org/10.1126/sciadv.aat3007.
- Olson, Wilma K, Manju Bansal, Stephen K Burley, Richard E Dickerson, Mark Gerstein, Stephen C Harvey, Udo Heinemann, et al. 2001. “A Standard Reference Frame for the Description of Nucleic Acid Base-Pair Geometry.” Journal of Molecular Biology 313 (1): 229–37. https://doi.org/10.1006/jmbi.2001.4987.

DNA and RNA are biological macromolecules consisting of long chains of nucleotides. In PDB coordinate files, each DNA/RNA chain is assigned a unique identifier. For the legacy PDB format, the size of the chain identifier is clearly defined to be one alphanumeric character. For the mmCIF format, the length of the chain identifier is flexible: it is normally up to 4-char long, but assembly files can have chain identifiers longer than 4 characters (as of May 2022, see examples).
Recently, I was approached with the following bug report where DSSR v2.4-2021nov11 was used:
Processing file '8feo-assembly1.cif'
[i] '8feo-assembly1.cif' taken as in .cif format by file extension.
*** buffer overflow detected ***: terminated
Aborted
I ran a newer version of DSSR (including the current release v2.5.2-2025apr03) on 8feo-assembly1.cif without any issue, as shown below:
# x3dna-dssr -i=8feo-assembly1.cif -o=8feo.out
[i] '8feo-assembly1.cif' taken as in .cif format by file extension.
Processing file '8feo-assembly1.cif'
[w] chain id 'AAA-2' > 4 chars
...
# Excerpt from 8feo.out
no. of DNA/RNA chains: 2 [AAA=16,AAA-2=16]
no. of nucleotides: 32
...
List of 16 base pairs
nt1 nt2 bp name Saenger LW DSSR
1 AAA.A1 AAA-2.U16 A-U WC 20-XX cWW cW-W
2 AAA.G2 AAA-2.C15 G-C WC 19-XIX cWW cW-W
3 AAA.A3 AAA-2.U14 A-U WC 20-XX cWW cW-W
The message [w] chain id 'AAA-2' > 4 chars is saying that the chain identifier ‘AAA-2’ is out of the 4-char limit.
In addition to 8feo, similar issues were also fixed for related PDB entries 5a79, 6a7a, 8feo, 8fep, 8feq, 7umk, and 4v3p. Note that 4v3p is a eukaryotic polyribosomal assembly which takes several hours to run on a MacBookPro with 32GB RAM.
Some background information on how DSSR handles chain identifiers for mmCIF format files
When mmCIF support was first added to DSSR in 2013, I hard-coded the chain identifier to 4 chars following the documentation. In early 2024, when running DSSR on weekly updated PDB entries, I noticed a core dump bug with PDB entry 8feo for its biological assembly 1. At that time, I was not aware of the update on mmCIF-Formatted Assembly Files and its expansion of chain identifiers for symmetry-related copies: with PDB assembly files, -# is appended to any chain that is generated by a symmetry operation. So if the base chain id has 3 chars (e.g., AAA), the symmetry related chain will have 5 chars (e.g., AAA-2).
That is the case for PDB entry 8feo: it has a chain with identifier AAA-2 which is symmetry-related to the asymmetric unit chain AAA. Since AAA-2 (5-char long) is above the hard-coded 4-char limit, DSSR crashed (out of array boundary in C). After recognizing the issue, I've increased the chain identifier limit in DSSR to 8 chars, more than enough for all current PDB entries. Moreover, DSSR performs sanity check of chain identifier length: it reports diagnostic message as shown above for chains with over 4-char identifiers (e.g., AAA-2), and automatically shortens long chains to the enlarged limit. DSSR is now more robust and user friendly: it no longer simply crashes, but communicates helpful info about unusual cases to draw users' attention.
Taking this opportunity, I have also proactively updated DSSR to support long atom names , residue names, and segment ids, in preparation for future id changes. Tracing issues to their root causes and fixing them systematically is a key part that makes DSSR a reliable tool for structural bioinformatics. Tests have been added to the quality control infrastructure to ensure that all these new features work as expected.
Nowadays, the vast majority (over 90%) of users’ questions about DSSR can be answered straight away simply because they have already been addressed in advance, as shown in the above example for long chain identifiers. I'm always on the lookout for issues reported on the 3DNA Forum, received from email, Zoom, or in person, and more systematically via DSSR update on weekly released PDB entries, and uploaded files on the web-services (g4.x3dna.org and skmatic.x3dna.org). Every issue is an opportunity to further polish DSSR and make it better. Overall, users’ feedback is invaluable to me: I take it as an asset instead of a burden.
Documentation on chain identifiers in PDB and mmCIF formats
PDB format
The Coordinate Section in the PDB format documentation contains the following for ATOM/HETATM records:
The ATOM records present the atomic coordinates for standard amino acids and nucleotides. ... Non-polymer chemical coordinates use the HETATM record type.
Record Format
COLUMNS DATA TYPE FIELD DEFINITION
-----------------------------------------------------------
22 Character chainID Chain identifier.
**Details**
- Non-blank alphanumerical character is used for chain identifier.
So the chain identifier in PDB format is a single alphanumeric character in column 22 of the ATOM/HETATM records.
mmCIF format
-
Large Structures Represented in mmCIF/PDBx:
Chain identifiers of up to 4 characters are permitted. The PDB chain identifier corresponds to the "_atom_site.auth_asym_id" data item.
-
News item on Distributing PDBx/mmCIF-Formatted Assembly Files
- Github repo on Sample assembly files in PDBx/mmCIF Format
These updated PDBx/mmCIF format assembly files files will include all symmetry generated copies of each chain within a single model, with distinct chain IDs assigned to each. Generation of distinct chain IDs in assembly files are based upon the following rules:
- Chain IDs of the original chains from the atomic coordinate file will be retained (e.g., A)
- Assign unique chain ID for each symmetry copy within a single model. Rules of chain ID assignments:
- The applied index of the symmetry operator will be appended to the original chain ID separated by a dash (e.g., A-2, A-3, etc.)
- If there are more than one type of symmetry operators applied to generate symmetry copy, a dash sign will be used between two operators (e.g., A-12-60, A-60-88, etc.)

Recently, I came across the paper by Mitra et al. (2025) titled "RNAproDB: A Webserver and Interactive Database for Analyzing Protein-RNA Interactions." I am glad to notice that DSSR (Lu et al. 2015) has been cited extensively in this work, as follows:
As part of the processing pipeline, multiple software is run including DSSR^12^ (base-pairing geometries, protein–RNA hydrogen bonds, and RNA secondary structure), HBPLUS^17^ (hydrogen bonds involving water molecules), ... Leontis-Westhof^27^ base pair annotations (as computed by DSSR^12^) ... The structural elements (stems, loops, hairpins, junctions, etc.) are detected using DSSR^12^ and mapped to the partial projection layout (via averaging corresponding residue coordinates)... We explored the relative abundance of different standard nucleotides (A, C, G, and U) in helical vs. non-helical regions (as computed by DSSR^12^)...We quantified the propensity of base-pairing (as detected by DSSR^12^) between different RNA bases (Fig. 3D).
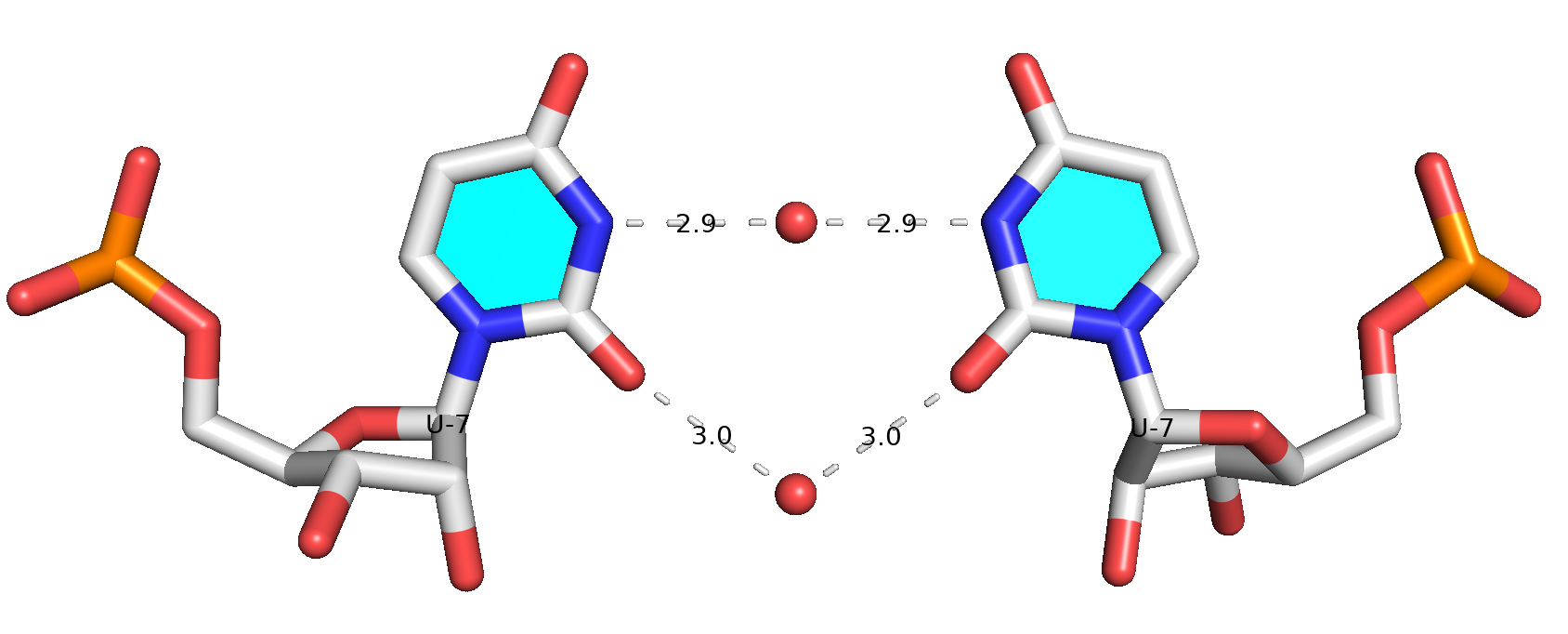
This is an impressive contribution on the characterization of protein-RNA interactions. Reading carefully through the paper and its supplemental PDF, I was intrigued by the following note on a water-mediated U-U base pair missed by DSSR.
Another important aspect to discuss is RNA–RNA water-mediated interactions^33,34^. ... One such example is the CUG repeat structure from PDB ID 7Y2B^35^ (Fig. S5A). The U/U mismatches in this structure are often unable to form direct hydrogen bonds (specifically, the central U/U mismatch forms no direct hydrogen bond). Therefore, DSSR^12^ does not classify it as a base pair. However, two water molecules form water-mediated hydrogen bonds between the two U bases. ...
While DSSR internally already takes consideration of water-mediated H-bonds in the detection of base pairs, it still requires: (1) at least one direct H-bond between two base atoms or a base atom to backbone, and (2) a co-planar geometry between the two bases. The water-mediated U7-U7 pair in PDB entry 7Y2B does not fulfill condition (1): the minimal distance between the two U bases is 5 Å, which is far larger than a typical H-bonding distance. Therefore, DSSR did not classify it as a base pair.
Prompted by the observation of Mitra et al. (2025), I have added a new option (--pair-water) in the DSSR v2.5.1-2025mar19 release to allow for water-mediated base pairs to be detected. Using PDB entry 7Y2B as an example, the DSSR command and related base-pairs output are shown below.
# x3dna-dssr -i=7Y2B.pdb1 --symm --pair-water
List of 13 base pairs
nt1 nt2 bp name Saenger LW DSSR
1 1:S.U1 2:S.A13 U-A WC 20-XX cWW cW-W
2 1:S.U2 2:S.A12 U-A WC 20-XX cWW cW-W
3 1:S.C3 2:S.G11 C-G WC 19-XIX cWW cW-W
4 1:S.U4 2:S.U10 U-U -- -- cWW cW-W
5 1:S.G5 2:S.C9 G-C WC 19-XIX cWW cW-W
6 1:S.C6 2:S.G8 C-G WC 19-XIX cWW cW-W
7 1:S.U7 2:S.U7 U-U Water -- cWW cW-W
8 1:S.G8 2:S.C6 G-C WC 19-XIX cWW cW-W
9 1:S.C9 2:S.G5 C-G WC 19-XIX cWW cW-W
10 1:S.U10 2:S.U4 U-U -- -- cWW cW-W
11 1:S.G11 2:S.C3 G-C WC 19-XIX cWW cW-W
12 1:S.A12 2:S.U2 A-U WC 20-XX cWW cW-W
13 1:S.A13 2:S.U1 A-U WC 20-XX cWW cW-W
Base pair #7 is water-mediated, as shown in the molecular image below. Note that .pdb1 means biological unit 1, and the option --symm reads the two symmetry-related structures in the MODEL/ENDMDL delineated ensemble as a single structure. See the DSSR User Manual for more details.
References
-
Lu, Xiang-Jun, Harmen J. Bussemaker, and Wilma K. Olson. 2015. “DSSR: An Integrated Software Tool for Dissecting the Spatial Structure of RNA.” Nucleic Acids Research, July, gkv716. https://doi.org/10.1093/nar/gkv716.
- Mitra, Raktim, Ari S. Cohen, Wei Yu Tang, Hirad Hosseini, Yongchan Hong, Helen M. Berman, and Remo Rohs. 2025. “RNAproDB: A Webserver and Interactive Database for Analyzing Protein-RNA Interactions.” Journal of Molecular Biology, February, 169012. https://doi.org/10.1016/j.jmb.2025.169012.









 "In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem." For
"In DSSR, a G4-helix is defined by stacking interactions of G-tetrads, regardless of backbone connectivity, and may contain more than one G4-stem." For  DSSR also reports the following loop:
DSSR also reports the following loop:

