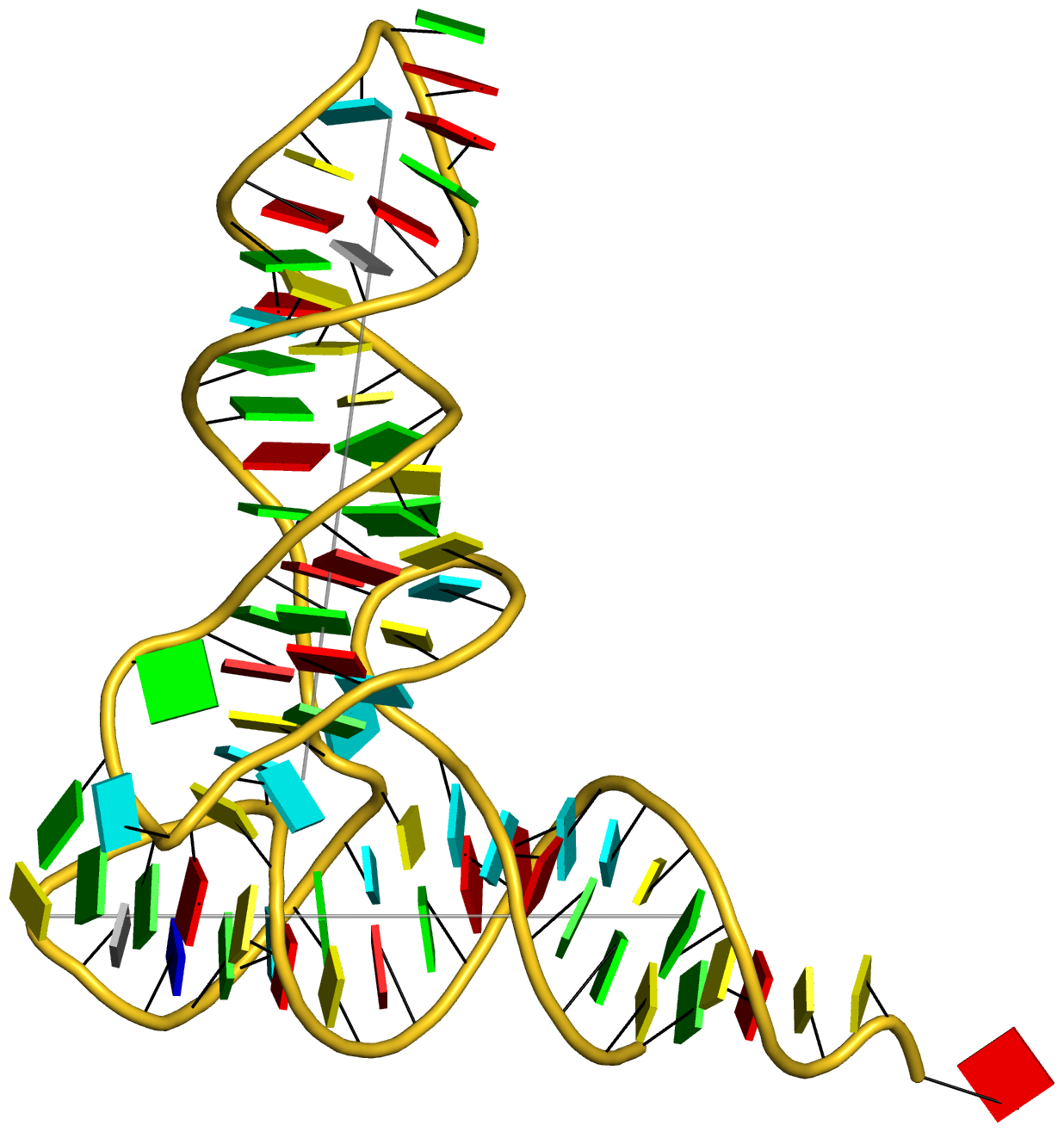
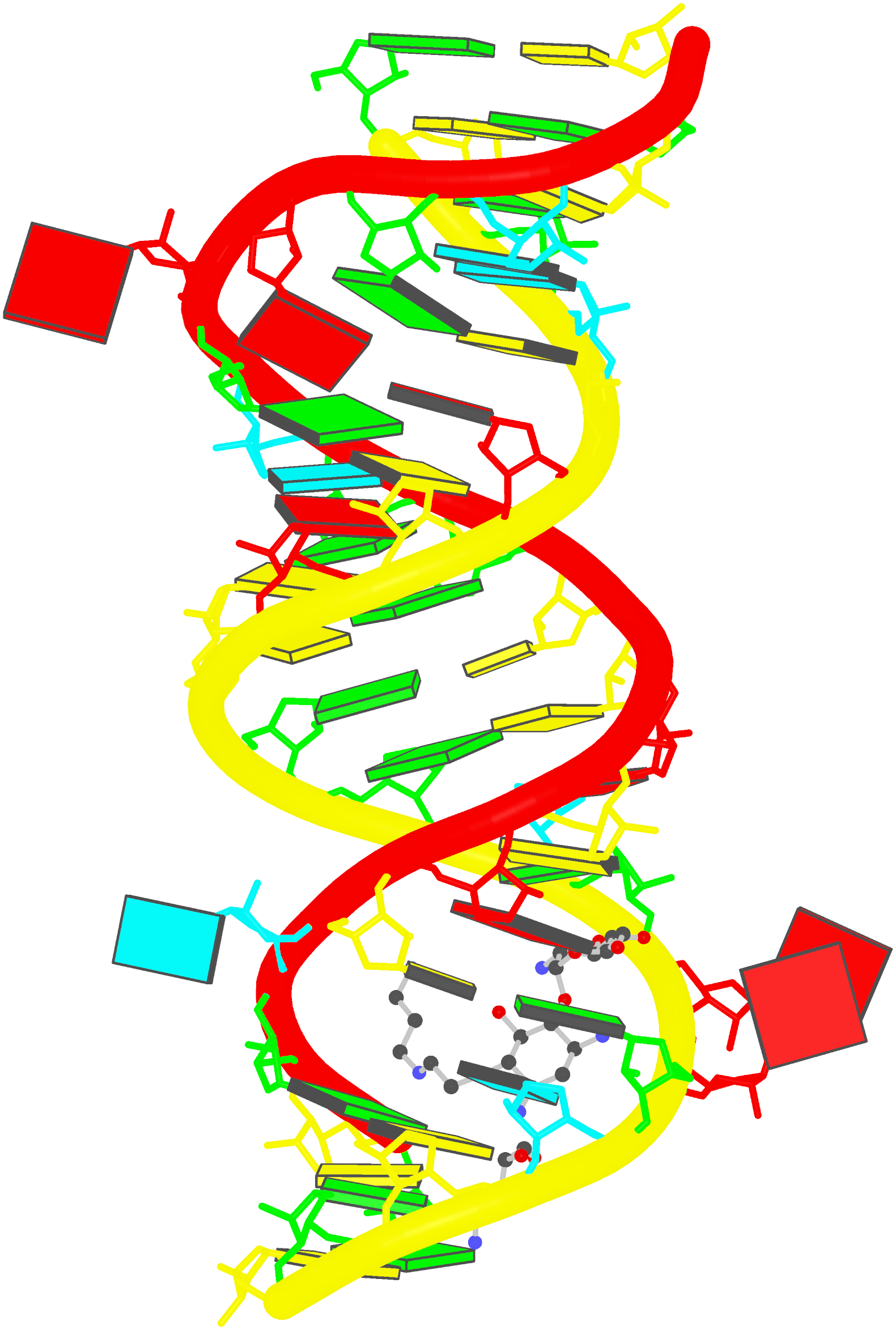
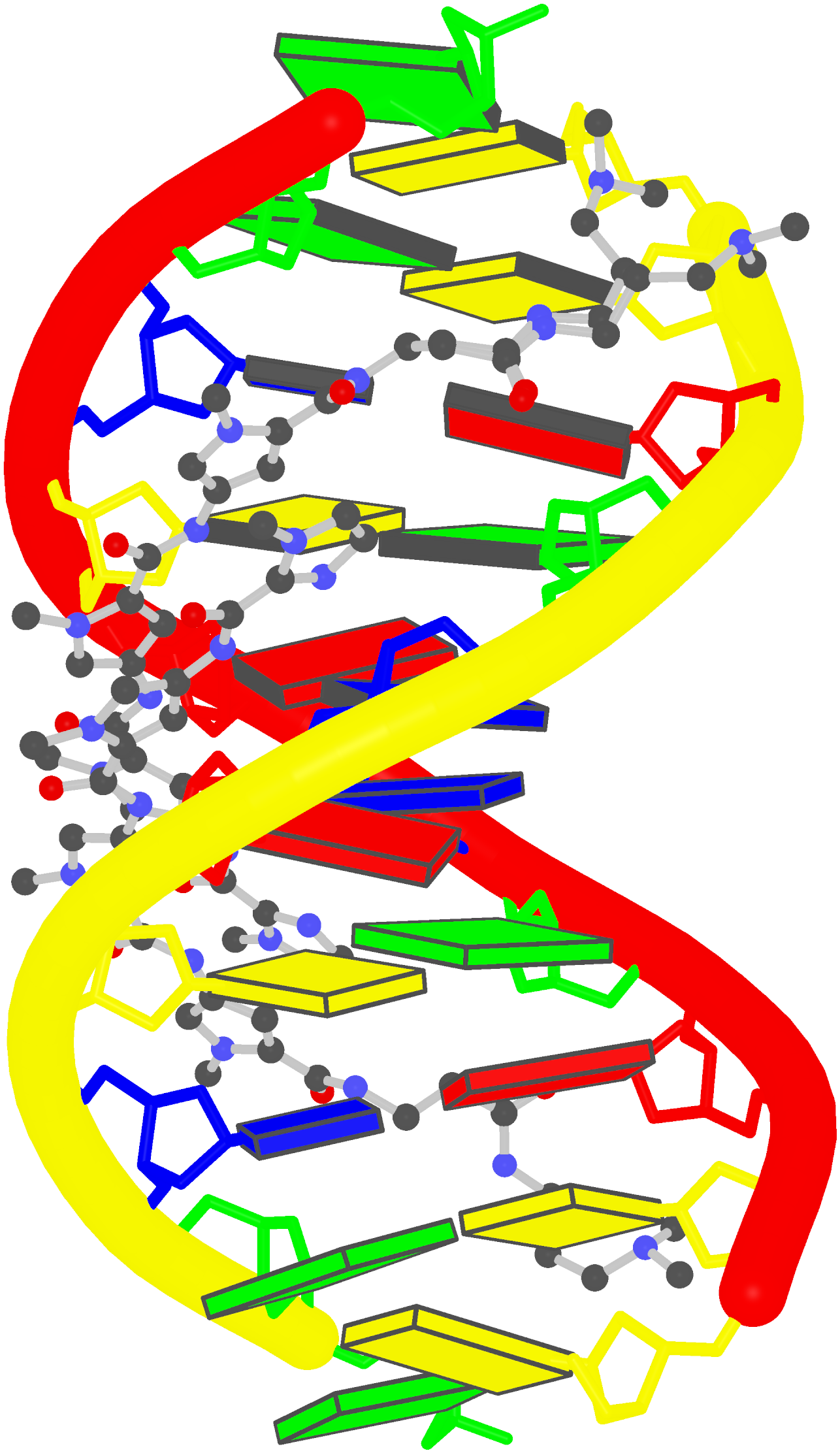
Prior to v2.1, 3DNA does not provide any direct support for the analysis of molecular dynamics (MD) simulations trajectories of nucleic acid structures. Nevertheless, over the years, I noticed some significant applications of 3DNA in the active MD field; see my blog post (December 6, 2009) titled 3DNA in the PCCP nucleic acid simulations themed issue. In January 2011, I released a set of two Ruby scripts specifically aimed to facilitate the analysis of MD simulations trajectories. Thereafter (as of 3DNA v2.1), I have significantly refined and expanded the Ruby scripts, and consolidated the functionality under one umbrella, x3dna_ensemble with multiple sub-commands (analyze, block_image, extract, and reorient). I believe x3dna_ensemble would make it straightforward to analyze ensembles (NMR or MD simulations trajectories) of nucleic acid structures.
Under this background, I am glad to read recently an article titled Structure, Stiffness and Substates of the Dickerson-Drew Dodecamer in J. Chem. Theory Comput. where 3DNA was used extensively. This work represents a re-visit of the classic Dickerson−Drew B-DNA dodecamer d-[CGCGAATTCGCG]2 using state-of-the-art MD simulations with different ionic conditions and solvation models, and compares the MD trajectories with modern crystallographic and NMR data. Among the author list (Tomas Drsata, Alberto Perez, Modesto Orozco, Alexandre Morozov, Jiri Sponer, and Filip Lankas) are some well-known figures in the MD field of nucleic acid structures.
Reading through the text, I am not sure if the newly available functionality of x3dna_ensemble was used. From the excerpts of the citations given below, however, it seems obvious that 3DNA is now well-accepted by the MD community.
Snapshots taken in 10 ps intervals were analyzed using the 3DNA program.43 From 3DNA outputs, time series of conformational parameters were extracted. These included the intra-base-pair coordinates (buckle, propeller, opening, shear, stretch, and stagger), inter-base-pair or step coordinates (tilt, roll, twist, shift, slide, and rise) as well as groove widths (based on P−P distances), backbone torsions, and sugar puckers.
Contrary to the original work of Lankas et al.,31 the intra-base-pair and step coordinates used here are those defined by 3DNA.43
Here, we apply this model together with the 3DNA definitions of the intra-base-pair and step coordinates.43
However, important differences remain, and non- negligible differences are in fact observed between individual experimental structures also in the central part of DD, even though the intra-base-pair and step coordinates are computed using the same coordinate definitions64 (we consistently use the 3DNA coordinates in this work).

Glycosidic bond “is a type of covalent bond that joins a carbohydrate (sugar) molecule to another group, which may or may not be another carbohydrate.” In nucleic acid structures, the other group is a nucleobase, and the predominated type is the N-glycosidic bond where the purine (A/G) N9 or pyrimidine (C/T/U) N1 atom connects to the C1′ atom of the five-membered (deoxy) ribose sugar ring. Another well-known type is the C-glycosidic bond in pseudouridine, the most common modified base in RNA structures where the C5 atom instead of N1 is linked to the C1′ atom of the sugar ring.

Recently, I performed a survey of all nucleic-acid-containing structures in the PDB/NDB database to see how many types of glycosidic bond are there. As always, I noticed some inconsistencies in the data: nucleotides with disconnected base/sugar, a base labeled as U but with pseudoU-type C-glycosidic bond. Shown below are a few unusual types of glycosidic bond in otherwise seemingly “normal” structures:
- The residue GN7 (number 28 on chain A) in PDB entry 1gn7 contains a N7-glycosylated guanine.
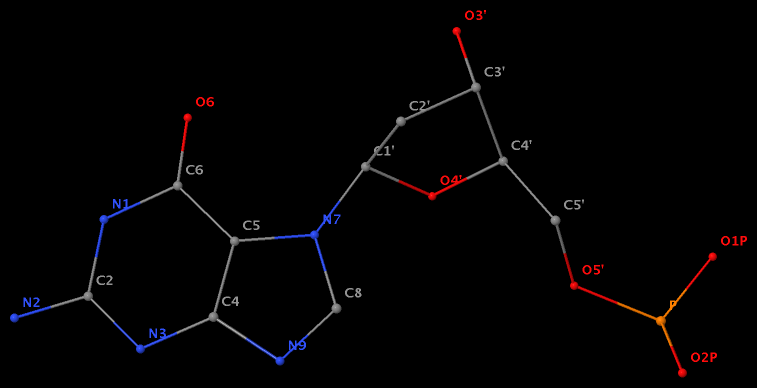
- The residue UPG (number 501 on chain A) in PDB entry 1y6f has sugar C1C (instead of C1′) atom connects to N1 of U.
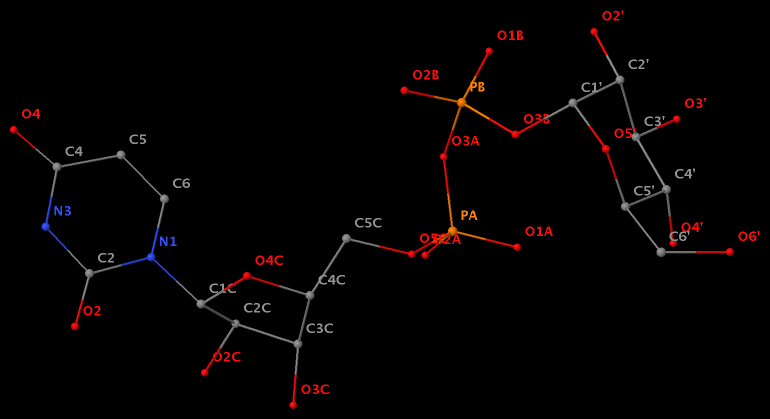
- The residue XAE (number 11 on chain B) in PDB entry 2icz contains a benzo-homologous adenine.
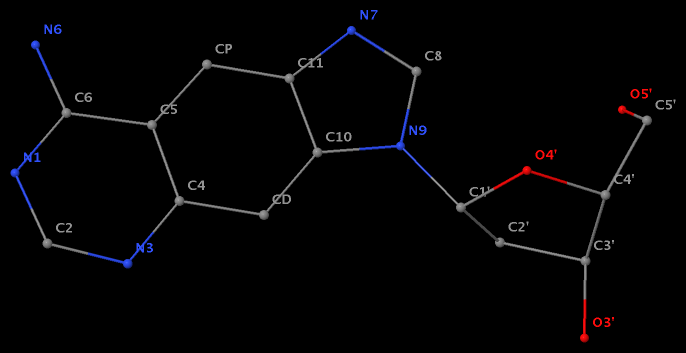
- The residue F5H (number 206 on chain B) in PDB entry 3v06 has N1 of U connects to C2′ of a six-membered sugar ring.
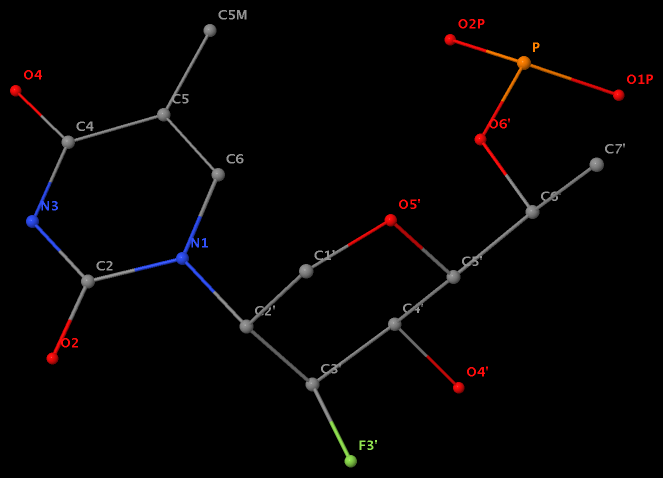
The unusual glycosidic bond has implications in 3DNA calculated parameters, for example the chi torsion angle. Identifying such cases would help refine 3DNA to provide sensible parameters and to avoid possible misinterpretations.

As of today (2012-09-16), the number of 3DNA forum registrations has reached 500! A quick browse of the ‘Statistics Center’ shows that over 80% of the registrations (400+) are after March 2012, when the new 3DNA homepage/forum were launched.
The sharp increase in registration is mostly due to the streamlined, web-based way to distribute the 3DNA software package. As far as I know, the number of 3DNA registrations/downloads in the past six months is significantly higher than that of 3DNA v2.0 for over three years. Equally importantly, I have been able to fixed every reported bug, addressed each feature request, and updated the 3DNA v2.1 distribution promptly.
I also feel confident to declare that up to now, the 3DNA Forum is spam free (at least to the extent I am aware). To this end, I’ve taken the following three measures:
- Installation of the SMF “Mod Stop Spammer”; as of this writing, it shows “3920 Spammers blocked up until today”.
- By using 3DNA-related verification questions. At its current setting, a user must answer correctly three of the ‘simple’ yet effective verification questions. Early on, I decided deliberately not to use CAPTCHA as an anti-spam means, based on my past experience.
- I’ve continuously monitored (new) registrations, and taken immediate actions against any suspicious registration. Due to the effectiveness of above two steps, so far I only have to manually handle just a few spam registrations. Nevertheless, it does illustrate the fact that no automatic method is perfect, and expert inspection is required to ensure desired results.
Overall, the new simplified way to distribute the 3DNA software package is working as intended; now users can easily access all distributed versions of 3DNA, and I can focus on support and further development of the software.

From a pure structural perspective, the designation of the two strands in an anti-parallel DNA duplex is sort of arbitrary. Thus, for a given PDB file, let’s assume that the atomic coordinates of chain A (strand I) come before those of chain B (strand II). We can swap the order of the two chains as they appear in the PDB file, i.e., list first the atomic coordinates of chain B and then those of chain A.
Structurally, the two settings corresponding to exactly the same DNA molecule. As far as 3DNA goes, however, the different orderings do make a different in calculated parameters. Using the Dickerson B-DNA dodecamer CGCGAATTCGCG solved at high resolution (PDB entry 355d) as an example, running 3DNA find_pair and analyze on ‘355d.pdb’ gives the results (abbreviated) below:
find_pair 355d.pdb 355d.bps
# contents of file '355d.bps':
------------------------------------------------------------------
355d.pdb
355d.out
2 # duplex
12 # number of base-pairs
1 1 # explicit bp numbering/hetero atoms
1 24 0 # 1 | ....>A:...1_:[.DC]C-----G[.DG]:..24_:B<....
2 23 0 # 2 | ....>A:...2_:[.DG]G-----C[.DC]:..23_:B<....
3 22 0 # 3 | ....>A:...3_:[.DC]C-----G[.DG]:..22_:B<....
4 21 0 # 4 | ....>A:...4_:[.DG]G-----C[.DC]:..21_:B<....
5 20 0 # 5 | ....>A:...5_:[.DA]A-----T[.DT]:..20_:B<....
6 19 0 # 6 | ....>A:...6_:[.DA]A-----T[.DT]:..19_:B<....
7 18 0 # 7 | ....>A:...7_:[.DT]T-----A[.DA]:..18_:B<....
8 17 0 # 8 | ....>A:...8_:[.DT]T-----A[.DA]:..17_:B<....
9 16 0 # 9 | ....>A:...9_:[.DC]C-----G[.DG]:..16_:B<....
10 15 0 # 10 | ....>A:..10_:[.DG]G-----C[.DC]:..15_:B<....
11 14 0 # 11 | ....>A:..11_:[.DC]C-----G[.DG]:..14_:B<....
12 13 0 # 12 | ....>A:..12_:[.DG]G-----C[.DC]:..13_:B<....
------------------------------------------------------------------
analyze 355d.bps
# generate output file '355d.out', with base-pair step parameters:
****************************************************************************
step Shift Slide Rise Tilt Roll Twist
1 CG/CG 0.09 0.04 3.20 -3.22 8.52 32.73
2 GC/GC 0.50 0.67 3.69 2.85 -9.06 43.88
3 CG/CG -0.14 0.59 3.00 0.97 11.30 25.11
4 GA/TC -0.45 -0.14 3.39 -1.59 1.37 37.50
5 AA/TT 0.17 -0.33 3.30 -0.33 0.46 37.52
6 AT/AT -0.01 -0.60 3.22 -0.31 -2.67 32.40
7 TT/AA -0.08 -0.40 3.22 1.68 -0.97 33.74
8 TC/GA -0.27 -0.23 3.47 0.68 -1.69 42.14
9 CG/CG 0.70 0.78 3.07 -3.66 4.18 26.58
10 GC/GC -1.31 0.36 3.37 -2.85 -9.37 41.60
11 CG/CG -0.31 0.21 3.17 -0.68 6.69 33.31
****************************************************************************
Reversing the order of chains A and B in ‘355d.pdb’ as ‘355d-reversed.pdb’ and repeating the above procedure, we have the following results:
find_pair 355d-reversed.pdb 355d-reversed.bps
# contents of file '355d-reversed.bps':
------------------------------------------------------------------
355d-reversed.pdb
355d-reversed.out
2 # duplex
12 # number of base-pairs
1 1 # explicit bp numbering/hetero atoms
1 24 0 # 1 | ....>B:..13_:[.DC]C-----G[.DG]:..12_:A<....
2 23 0 # 2 | ....>B:..14_:[.DG]G-----C[.DC]:..11_:A<....
3 22 0 # 3 | ....>B:..15_:[.DC]C-----G[.DG]:..10_:A<....
4 21 0 # 4 | ....>B:..16_:[.DG]G-----C[.DC]:...9_:A<....
5 20 0 # 5 | ....>B:..17_:[.DA]A-----T[.DT]:...8_:A<....
6 19 0 # 6 | ....>B:..18_:[.DA]A-----T[.DT]:...7_:A<....
7 18 0 # 7 | ....>B:..19_:[.DT]T-----A[.DA]:...6_:A<....
8 17 0 # 8 | ....>B:..20_:[.DT]T-----A[.DA]:...5_:A<....
9 16 0 # 9 | ....>B:..21_:[.DC]C-----G[.DG]:...4_:A<....
10 15 0 # 10 | ....>B:..22_:[.DG]G-----C[.DC]:...3_:A<....
11 14 0 # 11 | ....>B:..23_:[.DC]C-----G[.DG]:...2_:A<....
12 13 0 # 12 | ....>B:..24_:[.DG]G-----C[.DC]:...1_:A<....
------------------------------------------------------------------
analyze 355d-reversed.bps
# generate output file '355d-reversed.out', with base-pair step parameters:
****************************************************************************
step Shift Slide Rise Tilt Roll Twist
1 CG/CG 0.31 0.21 3.17 0.68 6.69 33.31
2 GC/GC 1.31 0.36 3.37 2.85 -9.37 41.60
3 CG/CG -0.70 0.78 3.07 3.66 4.18 26.58
4 GA/TC 0.27 -0.23 3.47 -0.68 -1.69 42.14
5 AA/TT 0.08 -0.40 3.22 -1.68 -0.97 33.74
6 AT/AT 0.01 -0.60 3.22 0.31 -2.67 32.40
7 TT/AA -0.17 -0.33 3.30 0.33 0.46 37.52
8 TC/GA 0.45 -0.14 3.39 1.59 1.37 37.50
9 CG/CG 0.14 0.59 3.00 -0.97 11.30 25.11
10 GC/GC -0.50 0.67 3.69 -2.85 -9.06 43.88
11 CG/CG -0.09 0.04 3.20 3.22 8.52 32.73
****************************************************************************
Comparing the base-pair step parameters between ‘355d.out’ and ’355d-reversed.out’, one would notice that while slide/rise/roll/twist simply switch orders, shift/tilt (the x-axis parameters) also flip their signs. On the other hand, the nucleotide serial numbers specifying base pairs (the left two columns) are identical in ‘355d.bps’ and ’355d-reversed.bps’.
Apart from explicitly swapping the two strands in PDB data file, one can simply switch around the nucleotide serial numbers generated with find_pair in order to analyze a DNA duplex based on its complementary sequence instead of the primary one. For example, starting from the same PDB file ‘355d.pdb’, we change ‘355d.bps’ to ’355d-cs.bps’ as below,
------------------------------------------------------------------
355d.pdb
355d-cs.out
2 # duplex
12 # number of base-pairs
1 1 # explicit bp numbering/hetero atoms
13 12
14 11
15 10
16 9
17 8
18 7
19 6
20 5
21 4
22 3
23 2
24 1
------------------------------------------------------------------
Run analyze 355d-cs.bps, one would get exactly the same parameters in output file ’355d-cs.out’ as in ’355d-reversed.out’.

As of v2.1, I’ve switched from Perl to Ruby as the scripting language for 3DNA. Consequently, the Perl scripts in previous versions of 3DNA (v1.5 and v2.0) are now obsolete. I’ll only correct bugs in existing Perl scripts, but will not add any new features.
For back reference, the scripts are still available from a separate directory $X3DNA/perl_scripts, with the following contents:
OP_Mxyz* dcmnfile* nmr_strs*
README del_ms* pdb_frag*
block_atom* expand_ids* x3dna2charmm_pdb*
blocview.pl* manalyze* x3dna_r3d2png*
bp_mutation* mstack2img* x3dna_setup.pl*
cp_std* nmr_ensemble* x3dna_utils.pm
Among them, x3dna_setup.pl and blocview.pl have corresponding Ruby versions: x3dna_setup and blocview. Actually, the .pl file extension (for Perl) was added to avoid confusion with the new Ruby scripts.
Some of the functionalities have been incorporated into the Ruby script x3dna_utils:
------------------------------------------------------------------------
A miscellaneous collection of 3DNA utilities
Usage: x3dna_utils [-h|-v] sub-command [-h] [options]
where sub-command must be one of:
block_atom -- generate a base block schematic representation
cp_std -- select standard PDB datasets for analyze/rebuild
dcmnfile -- remove fixed-name files generated with 3DNA
x3dna_r3d2png -- convert .r3d to image with Raster3D or PyMOL
------------------------------------------------------------------------
--version, -v: Print version and exit
--help, -h: Show this message
Along the same line, ensemble-related functionalities (for NMR or molecular dynamics simulations) have been consolidated and extended into the new Ruby script x3dna_ensemble:
------------------------------------------------------------------------
Utilities for the analysis and visualization of an ensemble
Usage: x3dna_ensemble [-h|-v] sub-command [-h] [options]
where sub-command must be one of:
analyze -- analyze MODEL/ENDMDL delineated ensemble (NMR or MD)
block_image -- generate a base block schematic image
extract -- extract structural parameters after running 'analyze'
reorient -- reorient models to a particular frame/orientation
------------------------------------------------------------------------
--version, -v: Print version and exit
--help, -h: Show this message
Conceivably, C programs in 3DNA can also be consolidated. For backward compatibility, however, all existing C programs will be kept — and refined as necessary — in the current 3DNA v2.x series. As of v3.x, I’ll completely re-organize 3DNA incorporating my years of experience in programming languages and knowledge of macromolecular structures.

In 3DNA, each base pair (bp) is specified by the identity of its two comprising nucleotides (nts), and their interactions. Some examples are shown below based on the PDB entry 1ehz (the crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution), with the shorthand form on the right:
....>A:...1_:[..G]G-----C[..C]:..72_:A<.... G-C
....>A:...4_:[..G]G-*---U[..U]:..69_:A<.... G-U
....>A:...9_:[..A]A-**+-A[..A]:..23_:A<.... A+A
....>A:..15_:[..G]G-**+-C[..C]:..48_:A<.... G+C
....>A:..26_:[M2G]g-**--A[..A]:..44_:A<.... g-A
Specification of a nucleotide
The nt specification string consists of 6 fields and follows the pattern below, with the number of characters in each field inside the parentheses:
modelNum(4)>chainId(1):ntNum(4)insCode(1):[ntName(3)]baseName(1)
- modelNum(4) — the model number is up to 4 digits, right-justified, with each leading space replaced by a dot. If no model number is available, as is the case for 1ehz (and virtually all other x-ray crystal structures in the PDB), it is written as
.... (4 dots).
- chainId(1) — the chain id is 1-char long, with space replaced by underscore.
- ntNum(4) — the nt residue number, handled as for the model number.
- insCode(1) — insertion code, handled as for the chain id.
- ntName(3) — the nt residue name is up to 3-char long, right-justified, with each leading space replaced by a dot.
- baseName(1) — the base name is 1-char long, mapped from ntName(3) following
$X3DNA/config/baselist.dat. Note that modified nucleotides are put in lower case to distinguish them from the canonical ones — for example, M2G to g.
For the complementary base in a bp, the order of the 6 fields is reversed — see examples above. To see the full list of nts in a PDB data file, run: find_pair -s 1ehz.pdb stdout (here using 1ehz as an example).
Specification of a base pair
The pattern of a bp is M-xyz-N, where M and N are 1-char base names (as in aforesaid field #6), and the three characters xyz have the following meaning:
z — the sign of the dot product of the z-axes of the M and N base reference frames. It is positive (+) if the two z-axes point in similar directions, as in Hoogsteen or reverse Watson-Crick bps. Conversely, it is negative (-) when the two z-axes point in opposite directions, as in the canonical Watson-Crick and Wobble bps. See figure below:

y — it is - if M and N are in a so-called Watson-Crick geometry (the two y-axes of the M and N base reference frames are anti-parallel, so are the two z-axes, whilst the two x-axes are parallel), e.g., the G-U Wobble pair; otherwise, *.x — it is - for Watson-Crick bps, otherwise, *.
By design, Watson-Crick bps would be of the pattern M-----N, Wobble bps M-*---N, and non-canonical bps M-**+-N or M-**--N. Thus by browsing through the 3DNA output, users can readily identify these three bp types.
The shortened form is represented as MzN; following aforementioned notation, it can be either M-N or M+N. The relative direction of the two z-axes is critical in effecting 3DNA-calculated bp (and step) parameters, as detailed in the 2003 3DNA NAR paper:
To calculate the six complementary base pair parameters of an M–N pair (Shear, Stretch, Stagger, Buckle, Propeller and Opening), where the two z‐axes run in opposite directions, the reference frame of the complementary base N is rotated about the x2‐axis by 180°, i.e. reversing the y2‐ and z2‐axes in Figure 2a. Under this convention, if the base pair is reckoned as an N–M pair, rather than an M–N pair, the x‐axis parameters (Shear and Buckle) reverse their signs. For an M+N pair, e.g. the Hoogsteen A+U in Figure 2b, the x2‐, y2‐ and z2‐axes do not change sign; thus all six parameters for an N+M pair are of opposite sign(s) from those for an M+N pair.
The M-N and M+N bp designation is unique to 3DNA. In combination with the corresponding 6 bp parameters (shear, stretch, stagger, buckle, propeller, and opening), 3DNA provides a rigorous description of all possible bps. This contrasts and complements with the conventional Saenger scheme and the 3-edge based Leontis/Westhof notation.
The 3DNA M-N vs M+N bp designation is base-centric, without concerning the sugar-phosphate backbone. The chi (χ) torsion angle, which characterizes base/sugar relative orientation, can be in either anti or syn conformation; thus similar backbone(S) can accommodate either M-N or M+N.

As the old saying goes, a picture is worth a thousand words. To help you have a better idea of what 3DNA/DSSR is about, we’ve collected the following pictures; they serve to demonstrate selected features from 3DNA/DSSR’s versatile functionality.
Schematic diagram of base-pair parameters
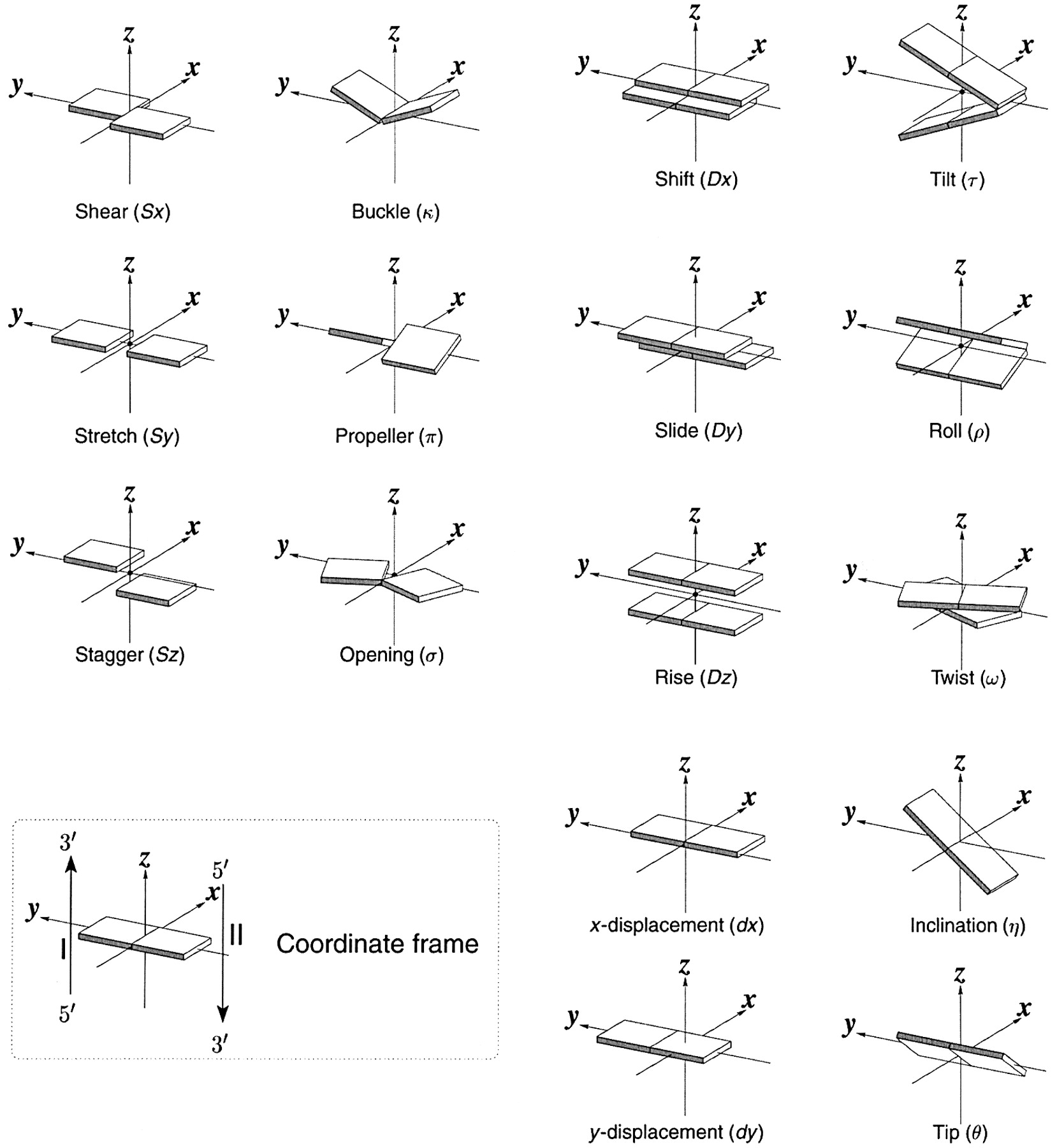
Influence of Slide and Roll on DNA helical conformation
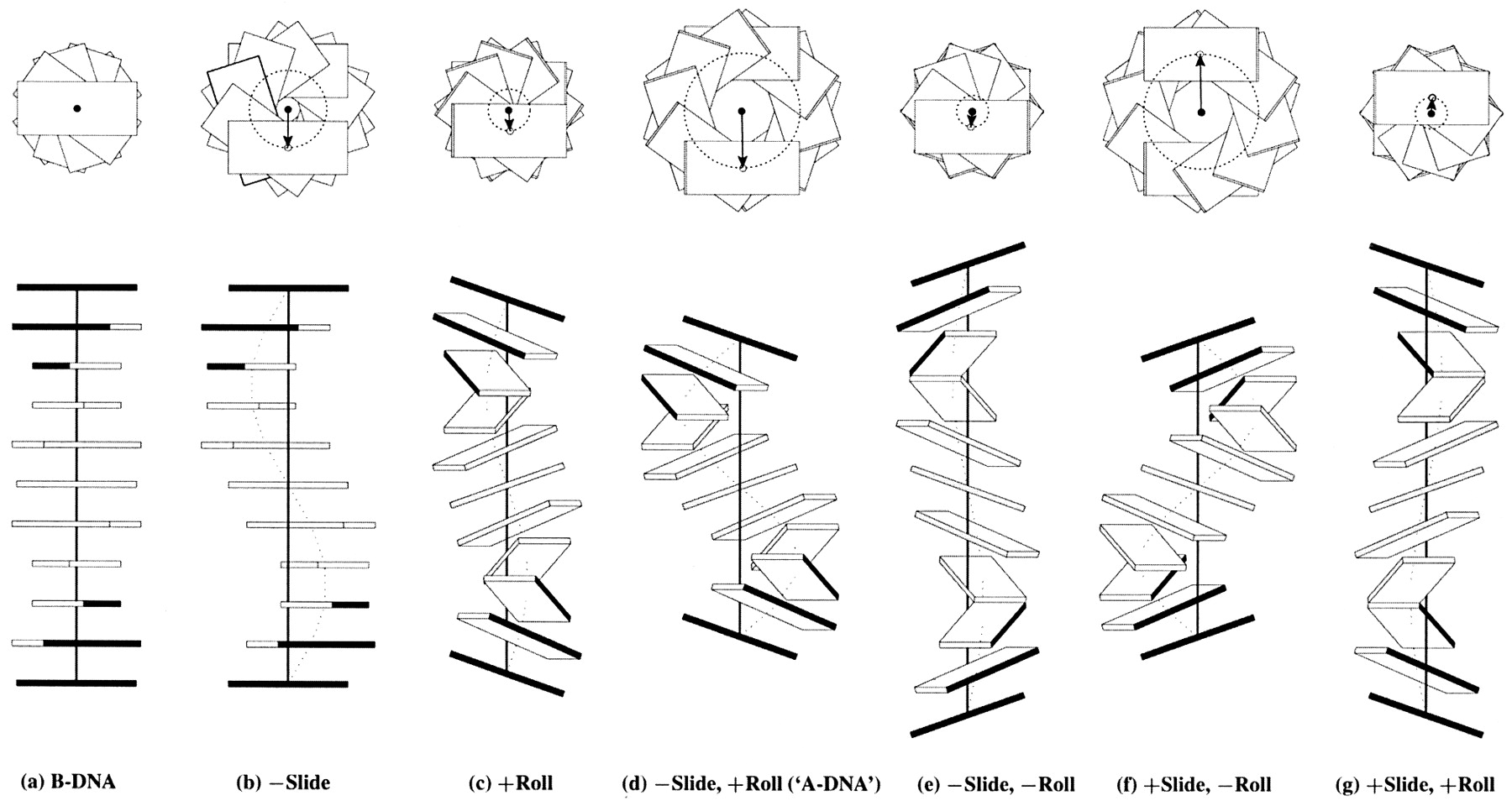
Roll-introduced DNA bending

Global bending of DNA associated with selective B → A conformational transformation

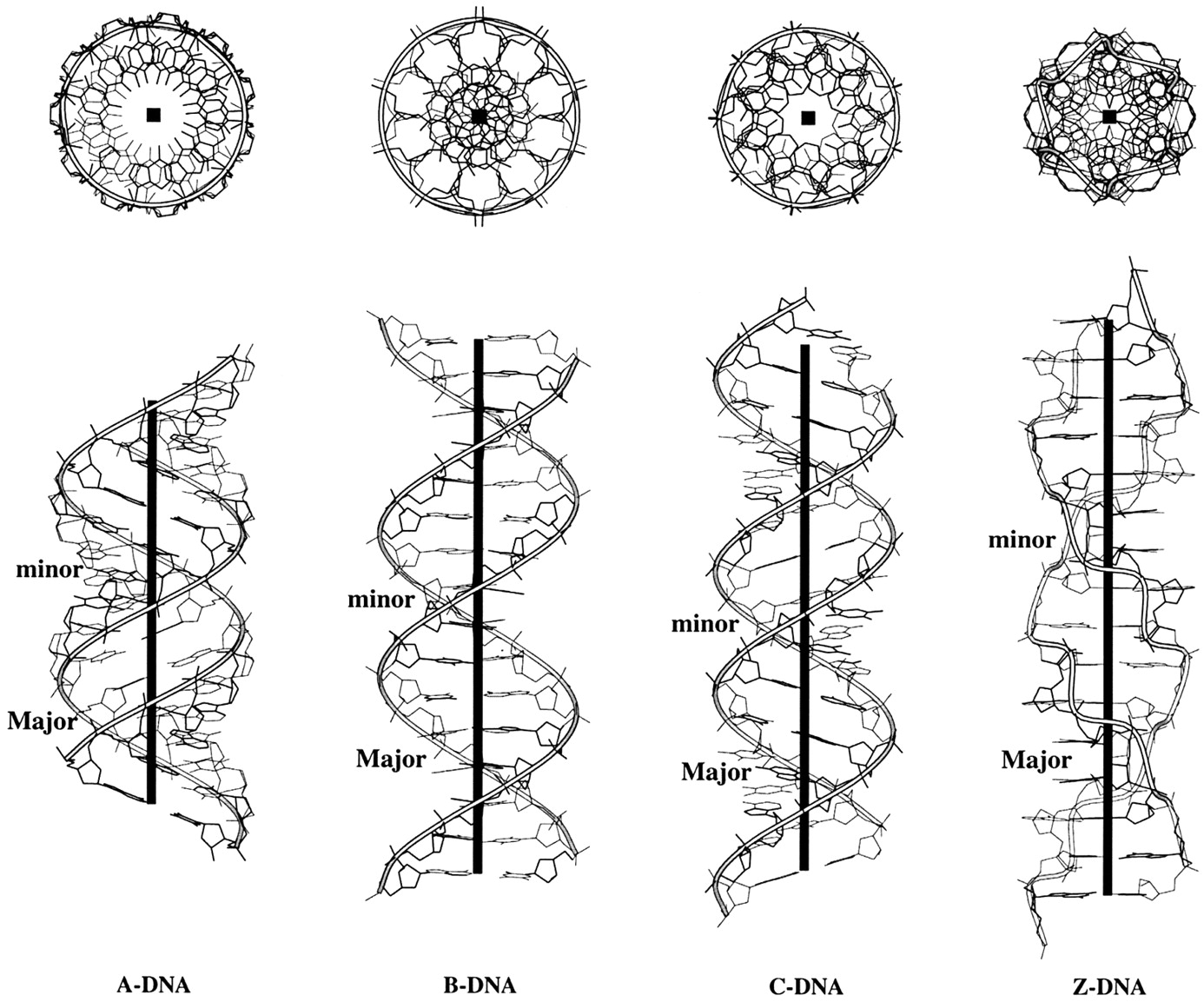
Canonical fiber models of A-, B-, C- and Z-DNA
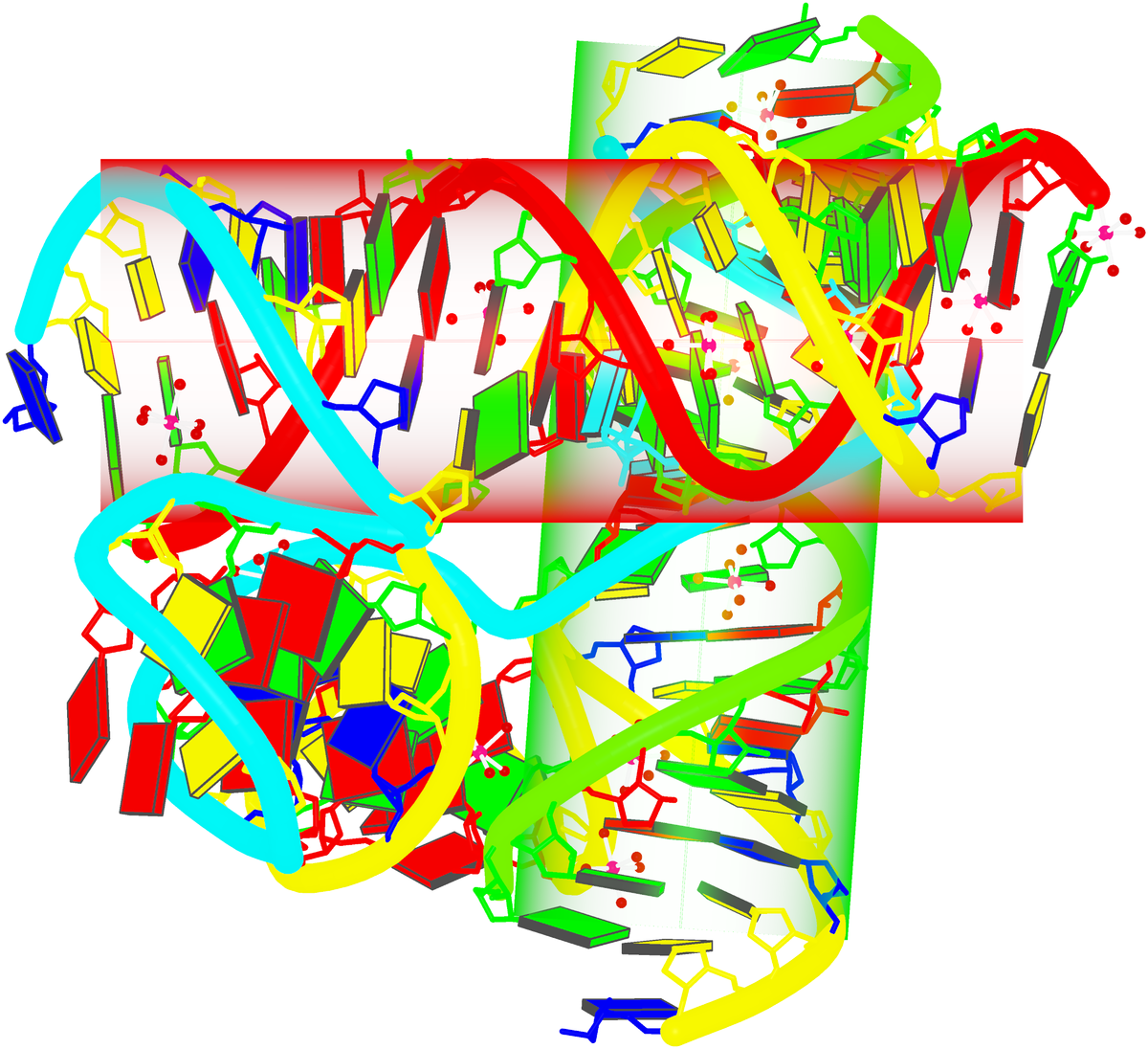
3DNA-generated view of a four-way DNA–RNA junction (1egk)
3DNA-detected pentaplets in the large ribosomal subunit (1jj2)
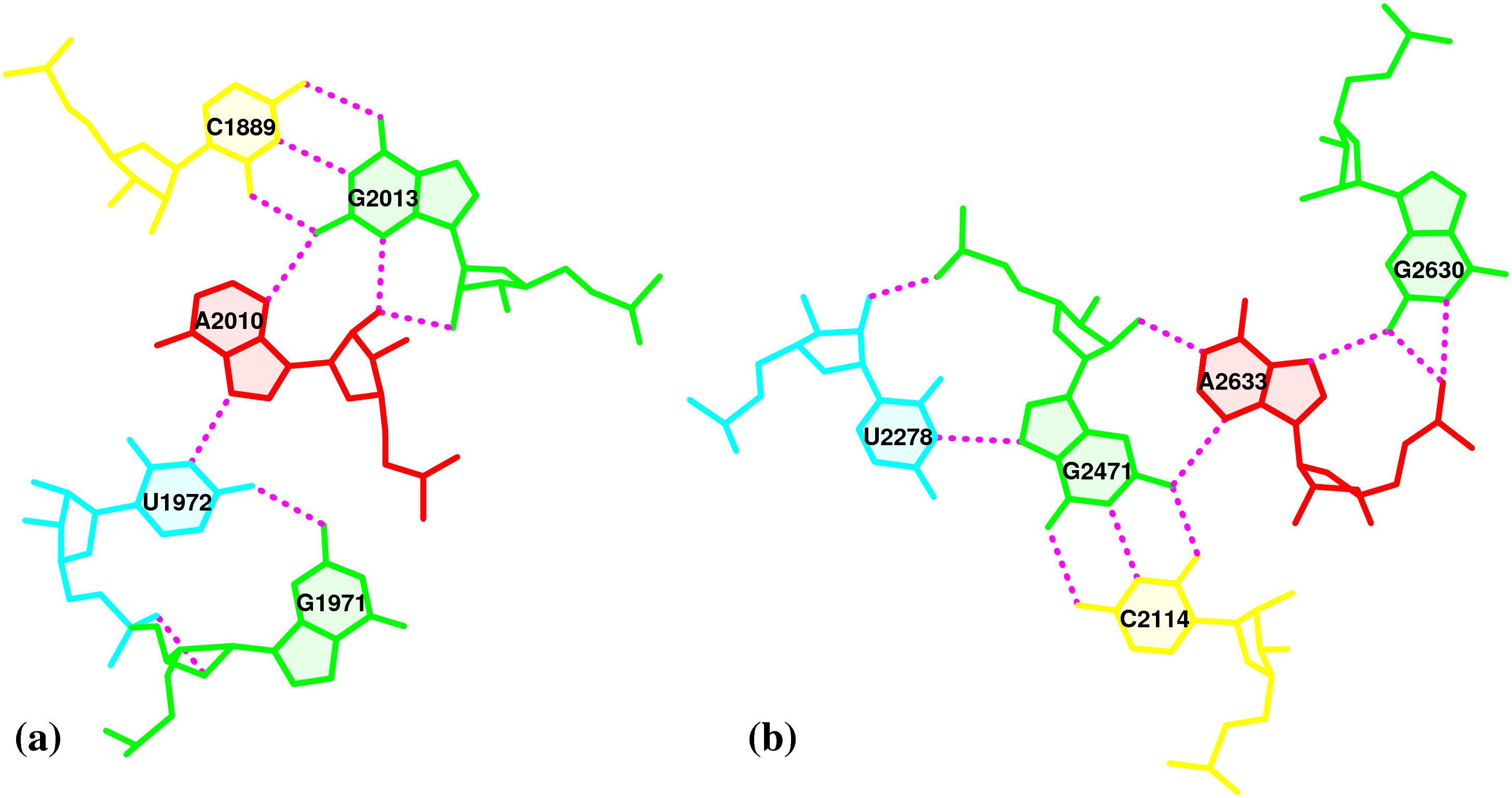
Nucleic-acid-containing structures generated with w3DNA
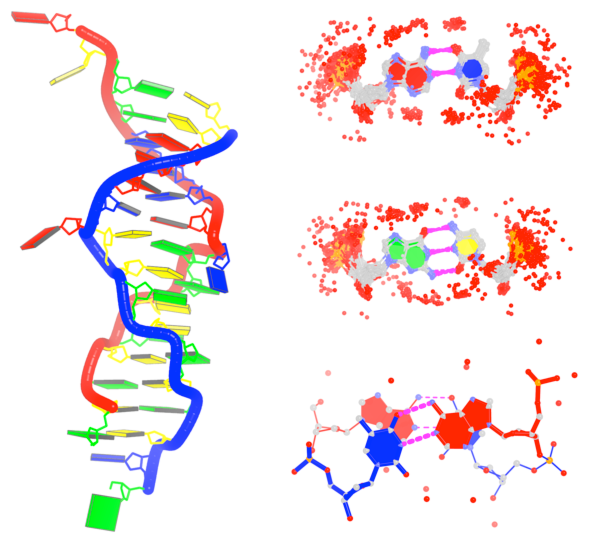
Analysis of DNA with a B-Z junction (2acj, left) and detection of hydration patterns (right)
Schematics images auto-generated via blocview

 A video overview of DSSR
A video overview of DSSR
DSSR (Dissecting the Spatial Structure of RNA) is an integrated software tool for the analysis/annotation, model building, and schematic visualization of 3D nucleic acid structures (see the figures below and the video overview). It is built upon the well-known, tested, and trusted 3DNA suite of programs. DSSR has been made possible by the developer’s extensive user-support experience, detail-oriented software engineering skills, and expert domain knowledge accumulated over two decades. It streamlines tasks in RNA/DNA structural bioinformatics, and outperforms its ‘competitors’ by far in terms of functionality, usability, and support.
Wide citations. DSSR has been widely cited in scientific literature, including: (i) “Selective small-molecule inhibition of an RNA structural element” (Nature, 2015; Merck Research Laboratories), (ii) “The structure of the yeast mitochondrial ribosome” (Science, 2017), (iii) “RNA force field with accuracy comparable to state-of-the-art protein force fields” (PNAS, 2018; D. E. Shaw Research), (iv) “Predicting site-binding modes of ions and water to nucleic acids using molecular solvation theory” (JACS, 2019), (v) “RIC-seq for global in situ profiling of RNA-RNA spatial interactions” (Nature, 2020), and (vi) “DNA mismatches reveal conformational penalties in protein-DNA recognition” (Nature, 2020).
Broad integrations. To make DSSR as widely accessible as possible, I have initiated collaborations with the principal developers of Jmol and PyMOL. The DSSR-Jmol and DSSR-PyMOL integrations bring unparalleled search capabilities (e.g., ‘select junctions’ for all multi-branch loops) and innovative visualization styles into 3D nucleic acid structures. DSSR has also been adopted into numerous other structural bioinformatics resources, including: (i) URS, (ii) RiboSketch, (iii) RNApdbee, (iv) forgi, (v) RNAvista, (vi) VeriNA3d, (vii) RNAMake, (viii) ElTetrado, (ix) DNAproDB, (x) LocalSTAR3D, (xi) IPANEMAP, and (xii) RNANet.
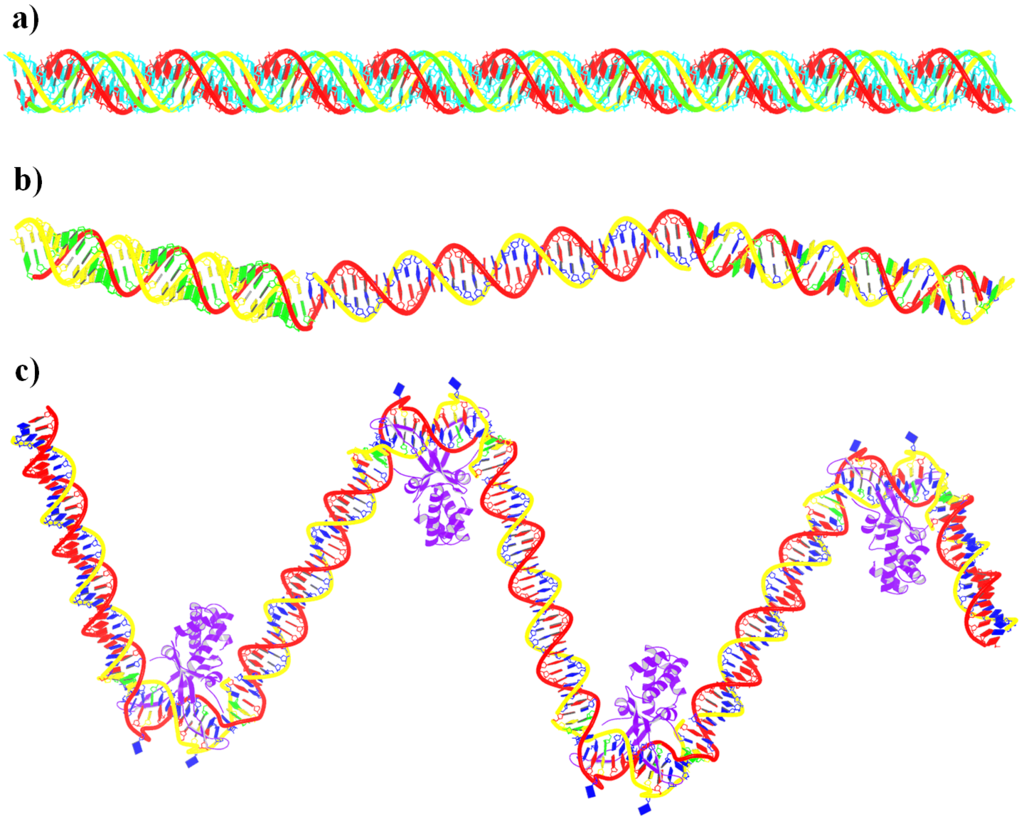
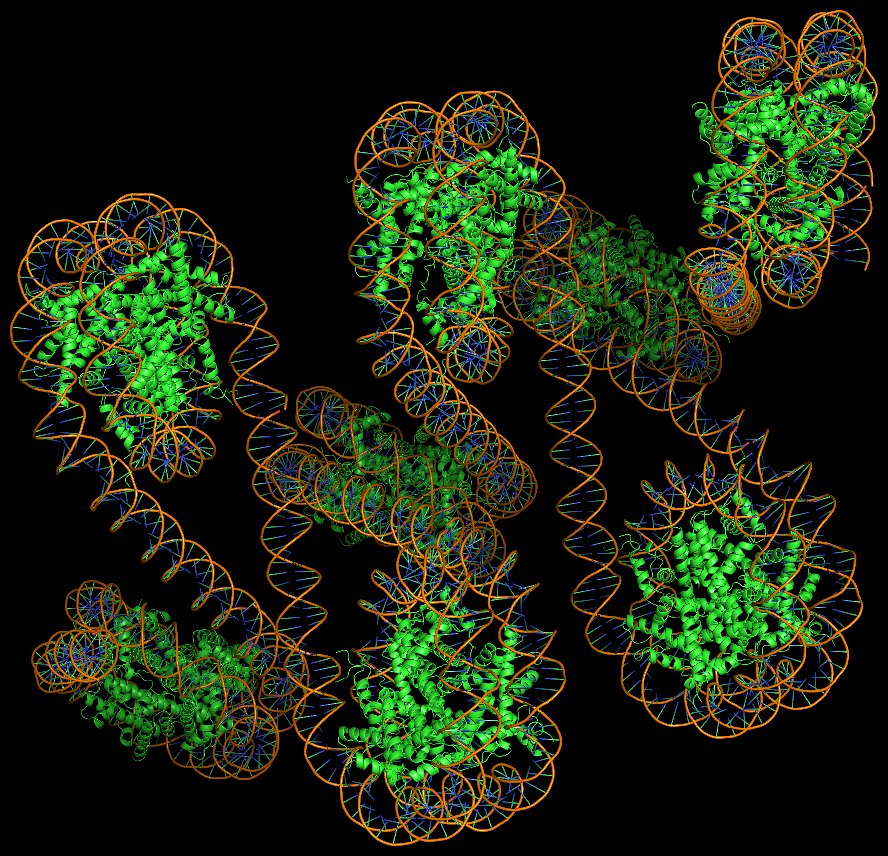
Advanced features. DSSR may be licensed from Columbia University. DSSR Pro is the commercial version. It has more functionalities than DSSR basic (the free academic version), including: (i) homology modeling via in silico base mutations, a feature employed by Merck scientists, (ii) easy generation of regular helical models, including circular or super-helical DNA (see figures below), (iii) creation of customized structures with user-specified base sequences and rigid-body parameters, (iv) efficient processing of molecular dynamics (MD) trajectories, (v) detailed characterization of DNA-protein or RNA-protein spatial interactions, and (vi) template-based modeling of DNA-protein complexes (see figures below). DSSR Pro supersedes 3DNA. It integrates the disparate analysis and modeling programs of 3DNA under one umbrella, and offers new advanced features, through a convenient interface. For example, with the mutate module of DSSR Pro, one can automatically perform the following tasks: (i) mutate all bases to Us, (ii) mutate bases in hairpin loops to Gs, and (iii) mutate G–C Watson-Crick pairs to C–G, and A–U to U–A. Moreover, DSSR Pro includes an in-depth user manual and one-year technical support from the developer.
Quality control. DSSR is a solid software product that excels in RNA structural bioinformatics. It is written in strict ANSI C, as a single command-line program. It is self-contained, with zero runtime dependencies on third-party libraries. The binary executables for macOS, Linux, and Windows are just ~2MB. DSSR has been extensively tested using all nucleic-acid-containing structures in the PDB. It is also routinely checked with Valgrind to avoid memory leaks. DSSR requires no set up or configuration: it simply works.

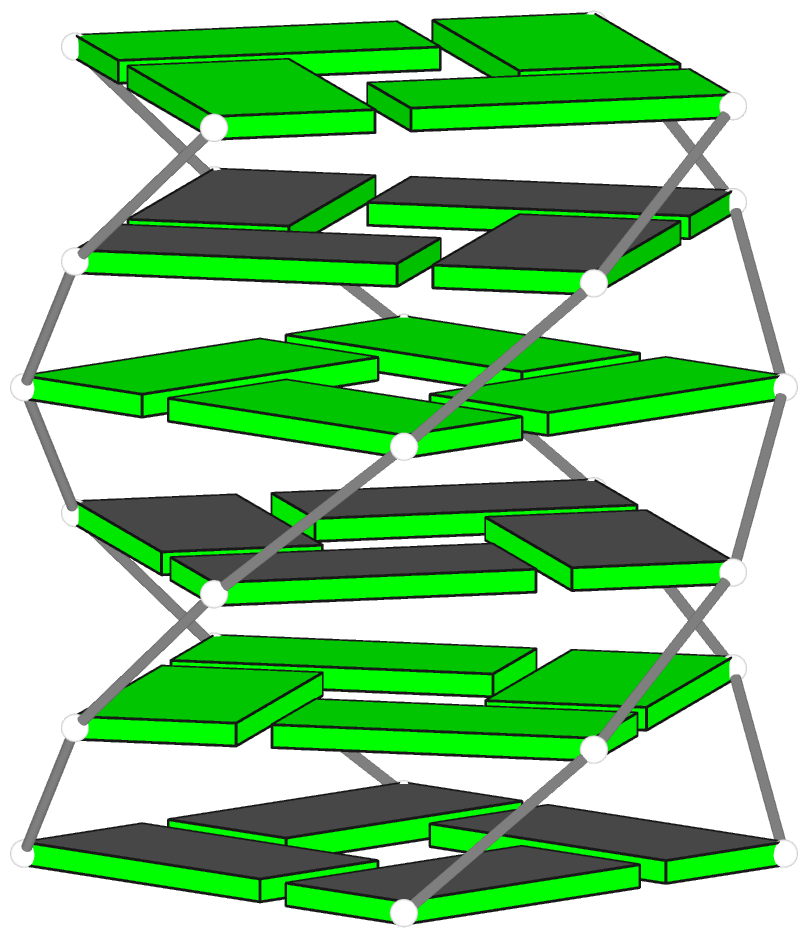
Theoretical models of G-quadruplexes, created using DSSR Pro.

Template-based modeling of DNA-protein complexes using DSSR Pro.
Here are two chromatin-like models using PDB entry 4xzq as the template.


Circular DNA duplexes modeled using DSSR Pro.
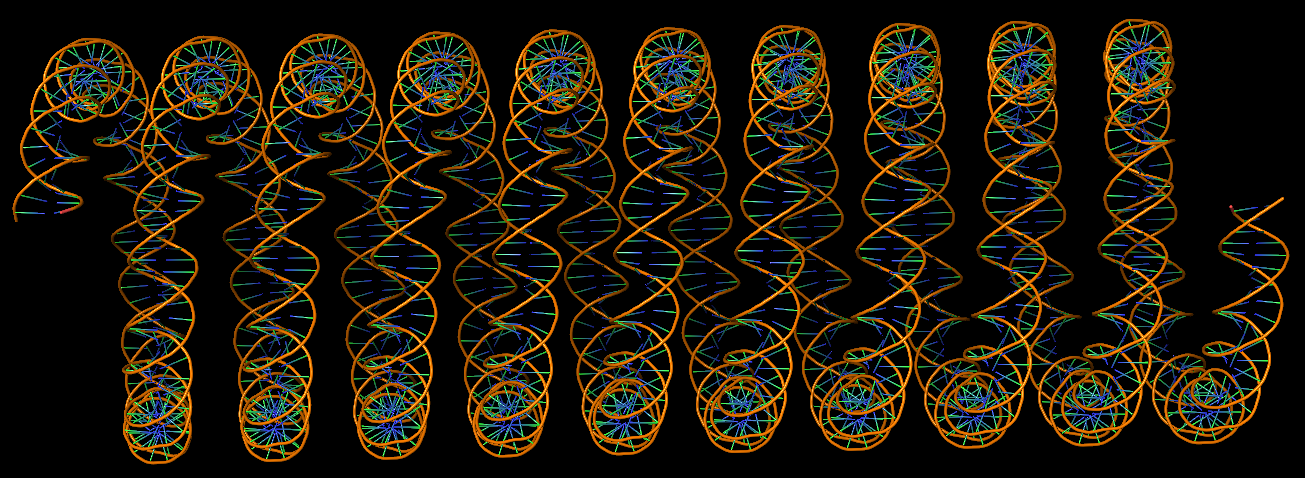
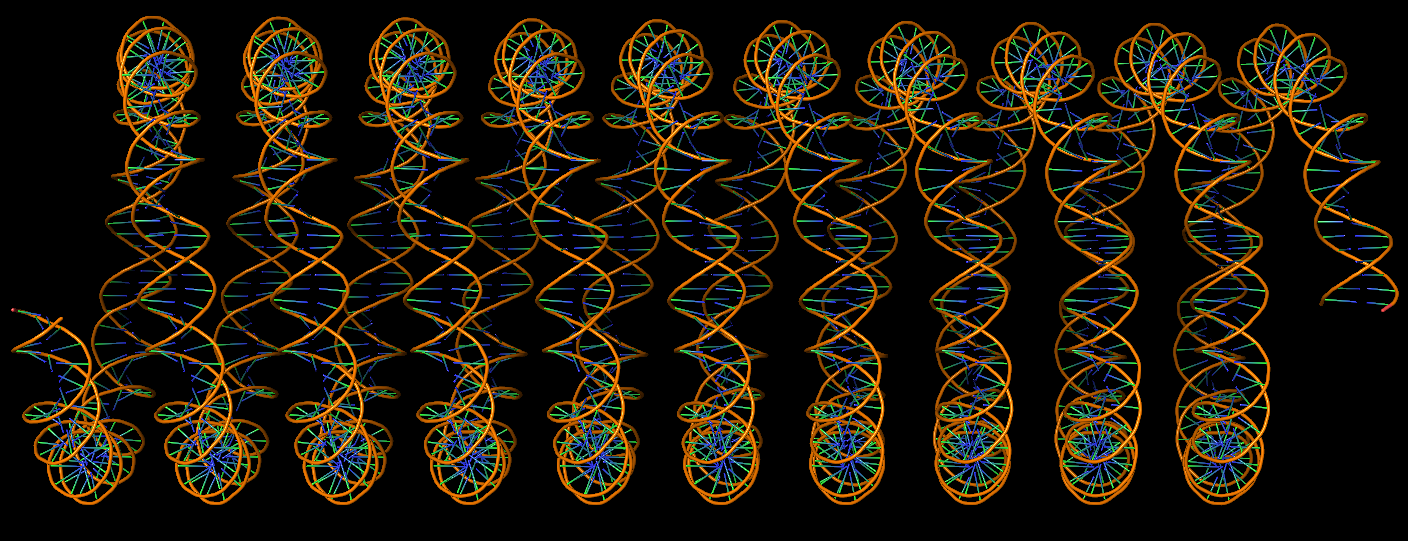
DNA super helices modeled using DSSR Pro.

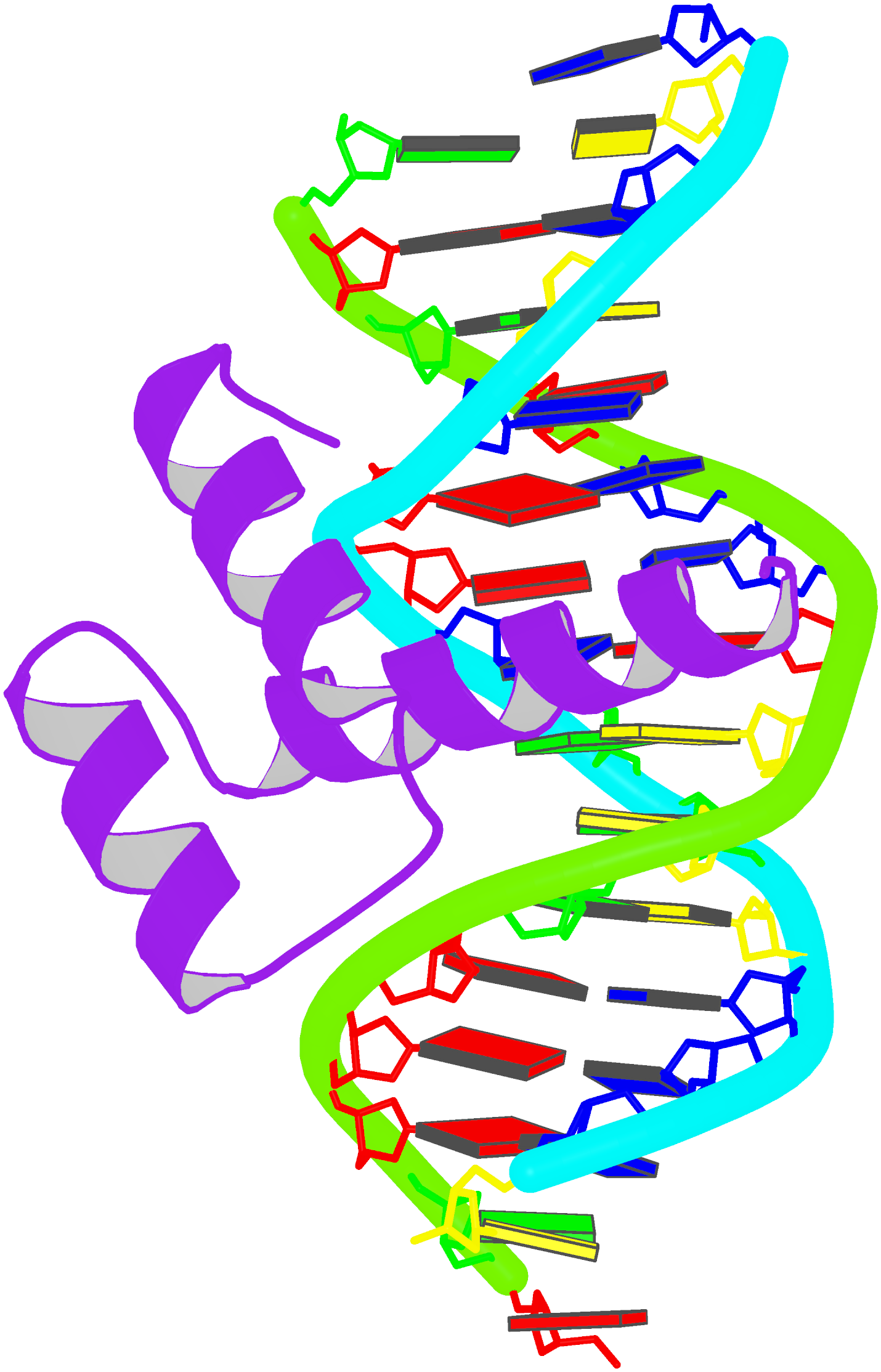
Innovative cartoon-block schematics enabled by the DSSR-PyMOL integration for six representative PDB entries. Watson-Crick pairs are shown as long blocks with minor-groove edges in black (A, B), G-tetrads represented as square blocks and the metal ion as sphere ©, the ligand rendered as balls-and-sticks (D), and proteins depicted as purple cartoons (E, F). Color code for base blocks: A, red; C, yellow; G, green; T, blue; U, cyan; G-tetrad, green; WC-pairs, per base in the leading strand. Visit http://skmatic.x3dna.org.
Recommended in Faculty Opinions: “simple and effective”, “Good for Teaching”.
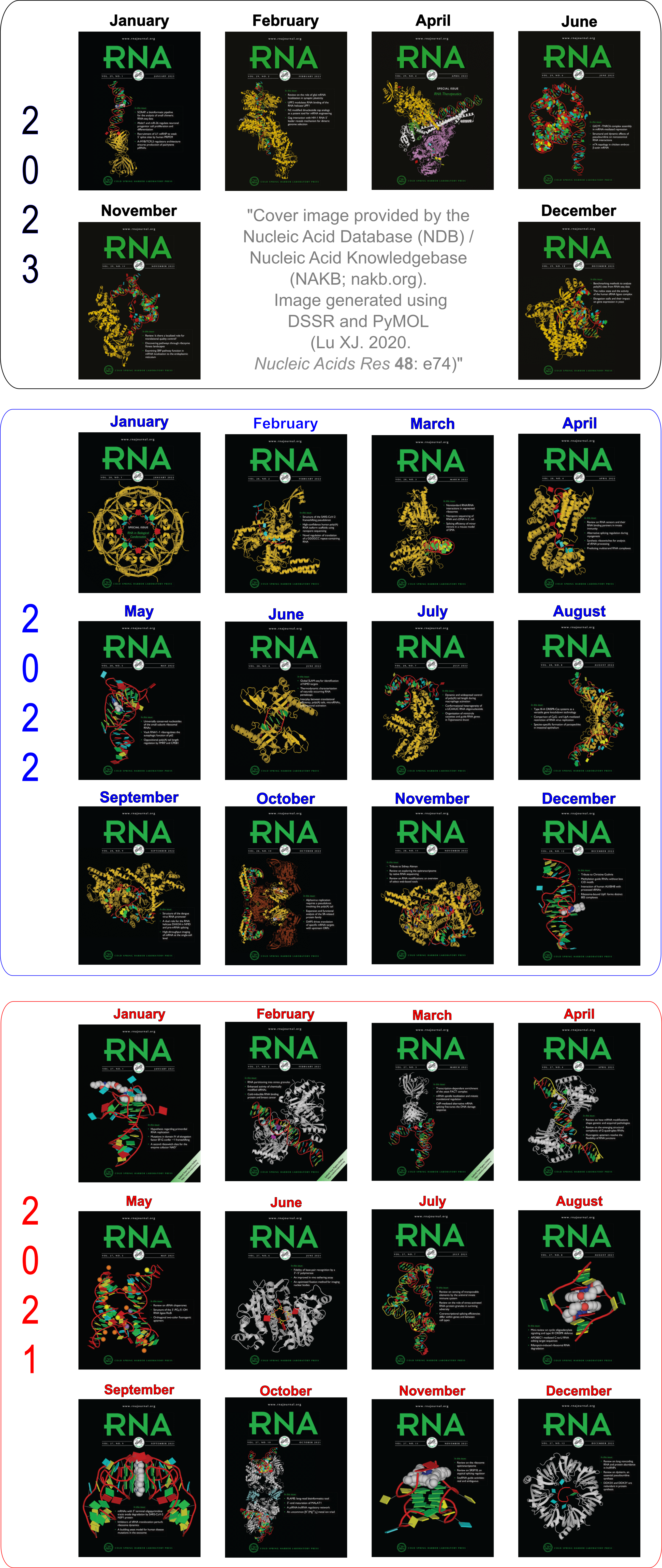
Employed by the NDB to create cover images of the RNA Journal.

The following links point to tools that are relevant to 3DNA.
- Curves+ — an updated version of the well-known Curves program, and it conforms to the standard base reference frame.
- 3D-DART — 3DNA-Driven DNA Analysis and Rebuilding Tool. Another web-interface to commonly used 3DNA functionality.
- do_x3dna — “do_x3dna has been developed for analysis of the DNA/RNA dynamics during the molecular dynamics simulations. It uses the 3DNA package to calculate several structural descriptors of DNA/RNA from the GROMACS MD trajectory. It executes 3DNA tools to calculate these descriptors and subsequently, extracts these output and saves into external output files as a function of time.”
- SwS — a Solvation web Service for Nucleic Acids where 3DNA plays a role.
- Raster3D — a set of tools for generating high-quality raster images of proteins or other molecules.
- MolScript — a program for displaying molecular 3D structures, such as proteins, in both schematic and detailed representations.
- Jmol — an open-source Java viewer for chemical structures in 3D with features for chemicals, crystals, materials, and biomolecules.
- PyMOL — a user-sponsored molecular visualization system on an open-source foundation.
- ImageMagick — a software suite to create, edit, compose, or convert bitmap images.
- NDB — Nucleic acids database.
- SBGrid — Excellent services for structural biology laboratories as well software developers.