Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. [Nucleic Acids Res 48: e74(https://doi.org/10.1093/nar/gkaa426)).
See the 2020 paper titled "DSSR-enabled innovative schematics of 3D nucleic acid structures with PyMOL" in Nucleic Acids Research and the corresponding Supplemental PDF for details. Many thanks to Drs. Wilma Olson and Cathy Lawson for their help in the preparation of the illustrations.
Details on how to reproduce the cover images are available on the 3DNA Forum.

Structure of a group II intron ribonucleoprotein in the pre-ligation state (PDB id: 8T2R; Xu L, Liu T, Chung K, Pyle AM. 2023. Structural insights into intron catalysis and dynamics during splicing. Nature 624: 682–688). The pre-ligation complex of the Agathobacter rectalis group II intron reverse transcriptase/maturase with intron and 5′-exon RNAs makes it possible to construct a picture of the splicing active site. The intron is depicted by a green ribbon, with bases and Watson-Crick base pairs represented as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; the 5′-exon is shown by white spheres and the protein by a gold ribbon. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Complex of terminal uridylyltransferase 7 (TUT7) with pre-miRNA and Lin28A (PDB id: 8OPT; Yi G, Ye M, Carrique L, El-Sagheer A, Brown T, Norbury CJ, Zhang P, Gilbert RJ. 2024. Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs. Nat Struct Mol Biol 31: 1426–1438). The RNA-binding pluripotency factor LIN28A invades and melts the RNA and affects the mechanism of action of the TUT7 enzyme. The RNA backbone is depicted by a red ribbon, with bases and Watson-Crick base pairs represented as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; TUT7 is represented by a gold ribbon and LIN28A by a white ribbon. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Cryo-EM structure of the pre-B complex (PDB id: 8QP8; Zhang Z, Kumar V, Dybkov O, Will CL, Zhong J, Ludwig SE, Urlaub H, Kastner B, Stark H, Lührmann R. 2024. Structural insights into the cross-exon to cross-intron spliceosome switch. Nature 630: 1012–1019). The pre-B complex is thought to be critical in the regulation of splicing reactions. Its structure suggests how the cross-exon and cross-intron spliceosome assembly pathways converge. The U4, U5, and U6 snRNA backbones are depicted respectively by blue, green, and red ribbons, with bases and Watson-Crick base pairs shown as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; the proteins are represented by gold ribbons. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Structure of the Hendra henipavirus (HeV) nucleoprotein (N) protein-RNA double-ring assembly (PDB id: 8C4H; Passchier TC, White JB, Maskell DP, Byrne MJ, Ranson NA, Edwards TA, Barr JN. 2024. The cryoEM structure of the Hendra henipavirus nucleoprotein reveals insights into paramyxoviral nucleocapsid architectures. Sci Rep 14: 14099). The HeV N protein adopts a bi-lobed fold, where the N- and C-terminal globular domains are bisected by an RNA binding cleft. Neighboring N proteins assemble laterally and completely encapsidate the viral genomic and antigenomic RNAs. The two RNAs are depicted by green and red ribbons. The U bases of the poly(U) model are shown as cyan blocks. Proteins are represented as semitransparent gold ribbons. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Structure of the helicase and C-terminal domains of Dicer-related helicase-1 (DRH-1) bound to dsRNA (PDB id: 8T5S; Consalvo CD, Aderounmu AM, Donelick HM, Aruscavage PJ, Eckert DM, Shen PS, Bass BL. 2024. Caenorhabditis elegans Dicer acts with the RIG-I-like helicase DRH-1 and RDE-4 to cleave dsRNA. eLife 13: RP93979. Cryo-EM structures of Dicer-1 in complex with DRH-1, RNAi deficient-4 (RDE-4), and dsRNA provide mechanistic insights into how these three proteins cooperate in antiviral defense. The dsRNA backbone is depicted by green and red ribbons. The U-A pairs of the poly(A)·poly(U) model are shown as long rectangular cyan blocks, with minor-groove edges colored white. The ADP ligand is represented by a red block and the protein by a gold ribbon. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).
Moreover, the following 30 [12(2021) + 12(2022) + 6(2023)] cover images of the RNA Journal were generated by the NAKB (nakb.org).
Cover image provided by the Nucleic Acid Database (NDB)/Nucleic Acid Knowledgebase (NAKB; nakb.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).
Aside from its extensive functionality for RNA structural analyses, DSSR also introduces a consistent and flexible way to process command-line options. Here, each option can be specified via a --key[=value] pair (or -key[=value] or key[=value]; i.e., two/one/zero preceding dashes are all accepted), key can be in either lower, UPPER or MiXed case, and value is optional for Boolean switches. Furthermore, options can be put in any order; if the same key is repeated more than once, the value specified last overwrites corresponding previous settings.
As always, the rules are best illustrated with concrete examples. Some typical use-cases are given below:
#1 analyze PDB entry '1msy', with default output to stdout
x3dna-dssr --input=1msy.pdb
#2 same as #1, with output directed to file '1msy.out'
x3dna-dssr --input=1msy.pdb --output=1msy.out
#3-6, same as #2
x3dna-dssr --output=1msy.out --input=1msy.pdb
x3dna-dssr --OUTPUT=1msy.out --Input=1msy.pdb
x3dna-dssr -output=1msy.out input=1msy.pdb
x3dna-dssr output=1msy.out --input=1msy.pdb
#7 the value '1ehz.pdb' overwrites '1msy.pdb'
x3dna-dssr --input=1msy.pdb input=1ehz.pdb
#8-12 with the switch --more set to true
x3dna-dssr -input=1msy.pdb --more
x3dna-dssr -input=1msy.pdb --more=true
x3dna-dssr -input=1msy.pdb --more=yes
x3dna-dssr -input=1msy.pdb --more=on
x3dna-dssr -input=1msy.pdb --more=1
#13 same as without specifying --more,
# or with values set to false/no/0
x3dna-dssr -input=1msy.pdb --more=off
#14 shorthand forms for --input and --output
x3dna-dssr -i=1msy.pdb -o=1msy.out
#15 it can also be more verbose
x3dna-dssr --input-pdb-file=1msy.pdb
#16-18 within a key, separator dash(-) and underscore (_)
# are treated the same, and can be omitted
x3dna-dssr -i=1msy.pdb -non-pair
x3dna-dssr -i=1msy.pdb -non_pair
x3dna-dssr -i=1msy.pdb -nonpair
By allowing for 2/1/0 dashes to precede each key and a dash/underscore character or none to separate words within the key, DSSR provides users with great flexibility in specifying command-line options to fit into their preferred styles. Not surprisingly, new programs to be added into 3DNA, or the version 3 release of the software will all follow the same convention.

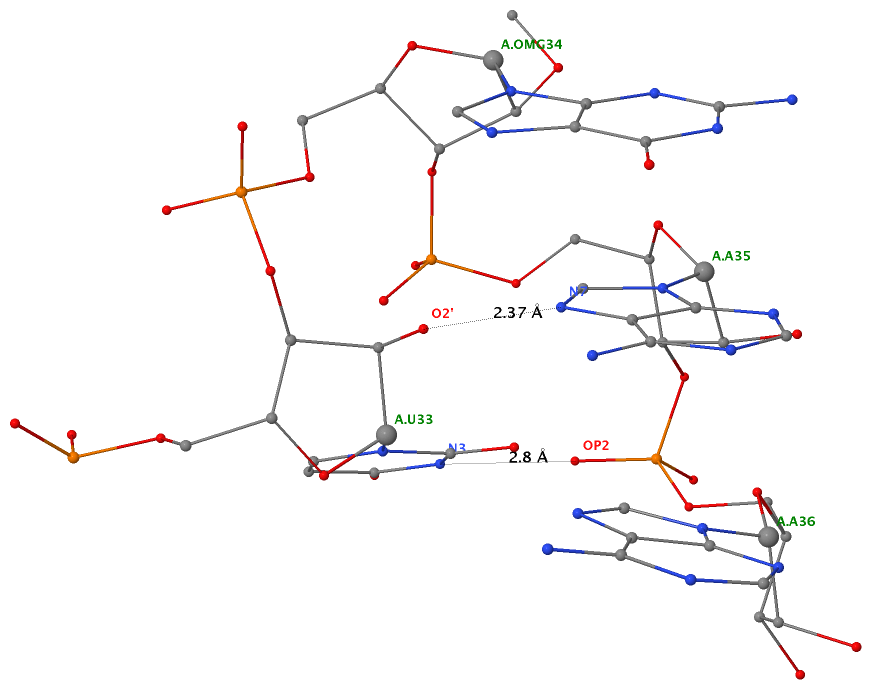
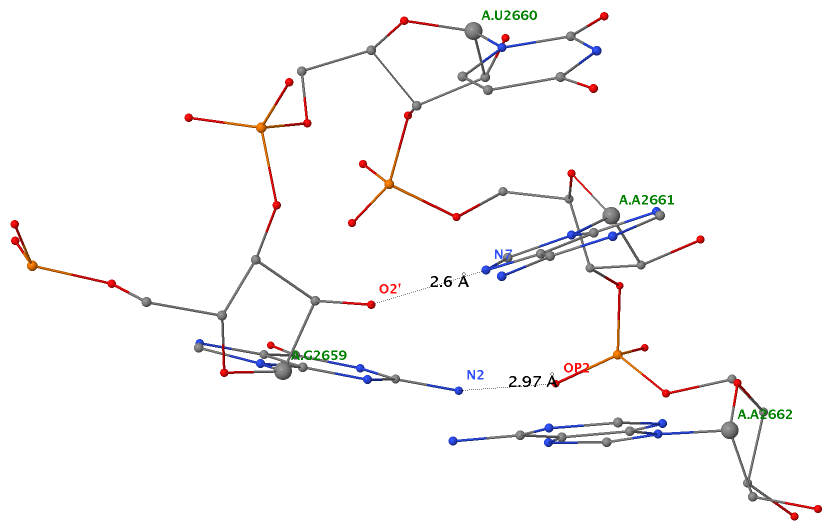
In addition to the five canonical bases (A, C, G, T, and U), nucleic acid structures in the PDB contains numerous modified variants (natural or engineered) in the nucleobase, sugar, or the phosphate. For instance, the 76-nt (nucleotide) long yeast phenylalanine tRNA (1ehz) contains 14 modified bases: 2MG10, H2U16, H2U17, M2G26, OMC32, OMG34, YYG37, PSU39, 5MC40, 7MG46, 5MC49, 5MU54, PSU55, and 1MA58. Among which, the most prevalent and best-known example is pseudouridine. Note that in the PDB, each residue (including modified nt) is named with an up to three-letter identifier, e.g., PSU for pseudouridine. For a comprehensive list (with chemical and structural information) of small molecules, including modified nts, please refer to the Ligand Expo website hosted by the RCSB PDB.
Given the widespread occurrences of modified bases in nucleic acid structures, any practical structural bioinformatics software should be able to treat them effectively, as with the canonical bases. In 3DNA, from the very beginning, modified bases are mapped to standard counterparts, e.g. 5‐iodouracil (5IU) to uracil (U) and 1‐methyladenine (1MA) to adenine (A), allowing for easy analysis of unusual DNA and RNA structures (see the NAR03 reference). Specifically, in the 3DNA distribution the file baselist.dat contains the mappings explicitly.
As of v2.1, 3DNA automatically maps a new modified base not available in the file baselist.dat. Yet, I have continuously updated the list in line with new DNA/RNA entries released by the PDB. The process is automated with a Ruby script which calls find_pair -s on each nucleic-acid-containing structure to output unknown bases. As an extreme, the baselist.dat file below comprises only canonical bases:
A A
C C
G G
T T
U U
DA A
DC C
DG G
DT T
With the above minimum mapping list, running the command find_pair -s on 1ehz.pdb identifies all the 14 modified bases. A sample case for 2MG is shown below:
Match '2MG' to 'g' for residue 2MG 10 on chain A [#10]
check it & consider to add line '2MG g' to file <baselist.dat>
By parsing the output of a batch run on all DNA/RNA-containing entries in the PDB as of October 18, 2013, I identified a total of 596 modified bases. The top portion is as below:
02I a
08Q c
08T a
0AD g
0C c
0DC c
0DG g
0DT t
0G g
0KL u
0KX c
0KZ t
An explicit list of base mapping makes the correspondence transparent, and helps avoid ambiguous cases as to which canonical base a modified nt matches to. DSSR uses the same list internally. Hopefully, the information would also be useful to other related projects.

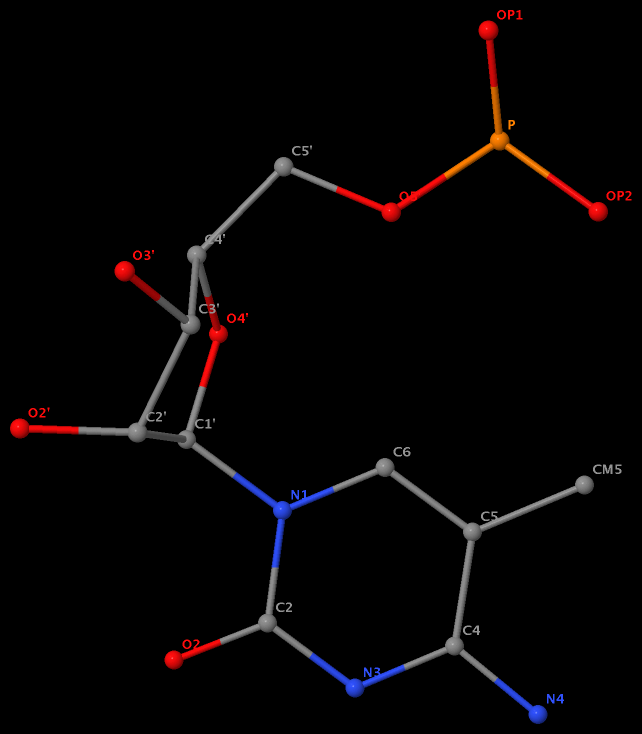
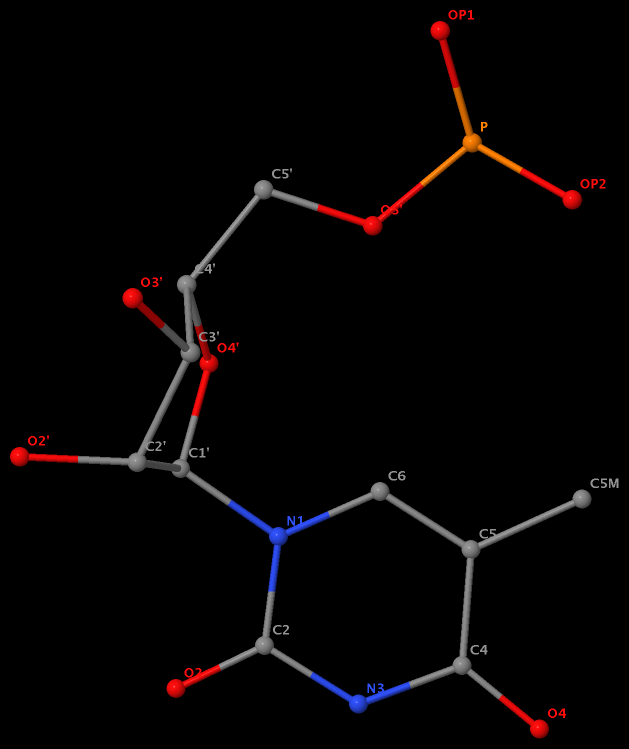
Recently I was a bit surprised to find that the methyl group is named differently in the PDB: C7 in DT8 (thymine) of B-DNA 355d, CM5 in 5MC40 (5-methylated C) of tRNA 1ehz, and C5M in 5MU54 (5-methylated U, i.e., T) of the same tRNA 1ehz. See the three figures below for details.
I know that the previously named C5M of thymine in DNA has been renamed C7 as a result of the 2007 remediation effort (PDB v3). However, browsing through the wwPDB Remediation website and reading carefully the article Remediation of the protein data bank archive, I failed to see explanations of the obvious inconsistency of CM5 (5MC40) vs C5M (5MU54) in the nomenclature of the 5-methyl group in the same tRNA entry 1ehz, except for the following note:
As with the Chemical Component Dictionary, names for standard amino acids and nucleotides follow IUPAC recommendations (10) with the exception of the well-established convention for C-terminal atoms OXT and HXT. These nomenclature changes have been applied to standard polymeric chemical components only.
5-methyl is named C7 in DT8 of the DNA entry 355d
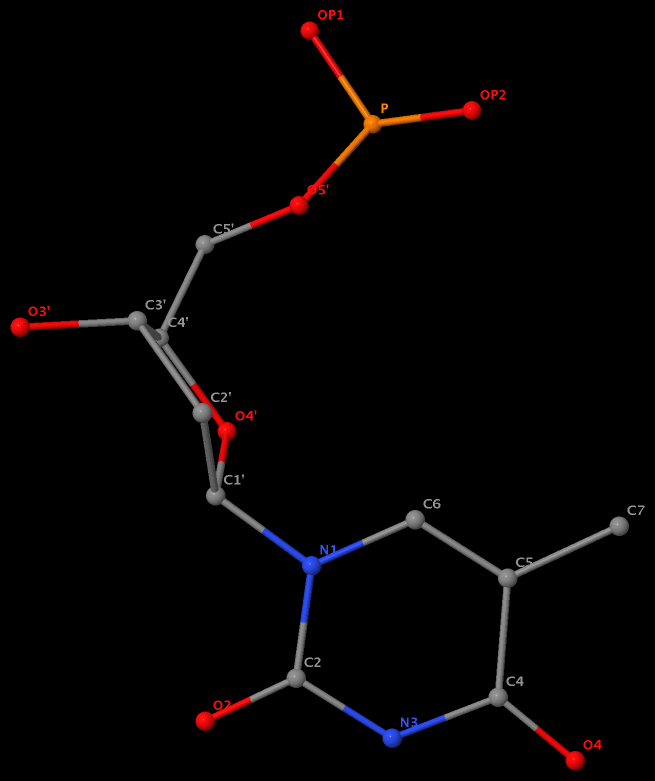
5-methyl is named CM5 in 5MC40 of the RNA entry 1ehz
5-methyl is named C5M in 5MU54 of the RNA entry 1ehz
Am I missing something obvious? If you have any further information, please leave a comment. Whatever the case, it helps (at least won’t hurt) to know the naming discrepancy for those who care about the small methyl group in nucleic acid structures.

Recently, I upgraded my local ViennaRNA package installation from v2.0.7 to v2.1.3 on my Mac. Following Quickstart in the INSTALL file, I ran ./configure successfully, but make aborted with error messages. Since I previously had a working copy of the software, it must be configuration issues when I compiled this new version. After a few iterations of checking the error message and reading through the INSTALL file, I came up with the following settings:
./configure --disable-openmp --without-perl
make
sudo make install
Apart from some warning messages, the above make command ran successfully.
This post serves mainly as a note for my own reference. Hopefully, the information may prove useful to others who try to install the versatile ViennaRNA package on a Mac OS X machine.

I’ve come up with a preliminary web-interface to DSSR, currently accessible at URL http://web.x3dna.org/dssr. The DSSR web-interface has been tested on Safari, Firefox, Chrome, and IE, with satisfying results. A screenshot of the home page is given below, using 1msy as an example:

After clicking the Submit button, users will be presented with the result page of a DSSR run. The beginning portion of the above example is as follows:

Note that the DSSR web-interface is being provided via a shared web hosting service, thus it has limited resources. Specifically, the uploaded file cannot be larger than two megabytes (2MB), and the process could be slow. Additionally, the file must have an extension of .pdb or .cif. To take full advantage of what DSSR has to offer, please install and run the software locally.
By design, DSSR is self-contained, command-line driven, with zero dependance on third-party libraries. Such features make it straightforward to build a GUI- or web-interface to DSSR, or integrate the program into other structural bioinformatics tools. As the need arises, I will refine the DSSR web-interface to better serve the community. The current simple, yet exploratory, web interface should make DSSR accessible to a much wider audience.

As of beta-r20-on-20130830, DSSR is able to detect two types of U-turns (see the figure below), the UNR-type (left) originally identified by Quigley and Rich [1976] in yeast phenylalanine tRNA, and the GNRA-type (right) later on established by Jucker and Pardi [1995] in GNRA tetra loops. See the Gutell et al. paper Predicting U-turns in Ribosomal RNA with Comparative Sequence Analysis for a more extensive account of U-turns.
As its name implies, a U-turn is characterized by a reversal of the RNA backbone direction within a few nucleotides. Among other factors, the U-turn is stabilized by two key H-bonding interactions, illustrated in dotted lines in the figure below.
Applying DSSR to 1jj2 (the crystal structure of the Haloarcula marismortui large ribosomal subunit) led to the identification of over 30 cases. In addition to the well-documented UNR- and GNRA-type U-turns, the program also finds other variants. An example is shown below, where the U-turn is within a GCA triloop instead of a GNRA tetraloop. Here, the N1 (not N2) atom of G1809 forms an H-bond with OP2 of G1812. The G1809 N2 atom is H-bonded to G1812 O5′ to further stabilize the U-turn.
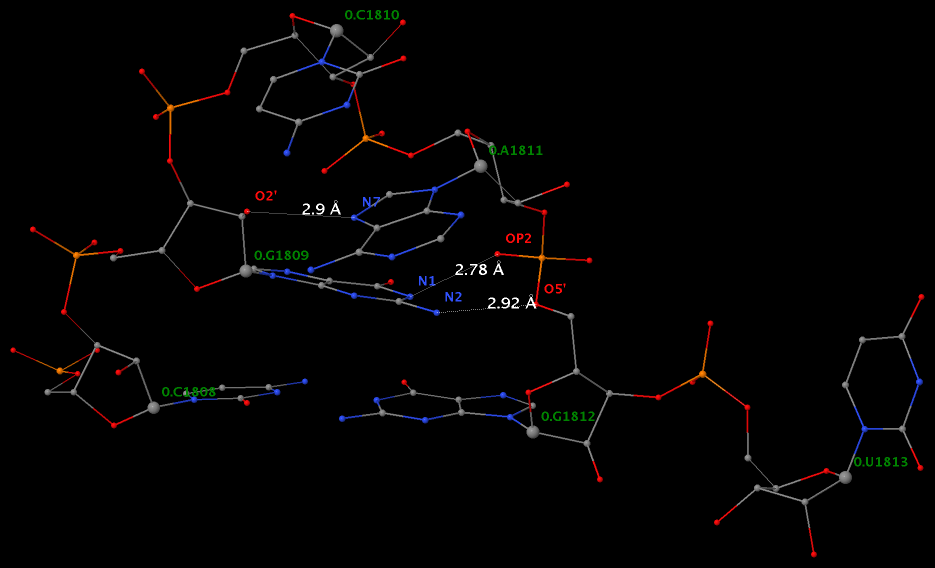
An examination of the chemical structure of the nitrogenous bases (see figure below) shows clearly other possibilities to connect RNA base donors to the phosphate oxygen acceptors. DSSR allows for the exploration of such variations, and more.


3DNA can build DNA/RNA structures with a precise base but approximate sugar-phosphate backbone geometry. In the 2003 3DNA-NAR paper, Table 3 of the section “Structures built with sugar–phosphate backbone” lists “root mean square deviation (in Å) between rebuilt 3DNA models and experimental DNA structures” for three representative DNA structures (in A-form, B-form, and a protein-DNA complex). It was noted that The RMSD of reconstructed versus observed base positions is virtually zero and that for both base and backbone coordinates is <0.85 Å, even for the 146 bp nucleosomal DNA structure.
The backbone geometry is approximate because 3DNA uses a fixed sugar-phosphate conformation (in A-DNA, B-DNA or RNA) that is attached to the corresponding bases in the model building process. The most noticeable effect is the long O3′(i)···P(i+1) bond that connects consecutive nucleotides along a chain. The imprecise structure was intended as a starting point for other objectives (e.g., all-atom molecular dynamics simulations) that are out of the design scope of 3DNA. Nevertheless, over the years, I have been concerned with the overlong O3′—P distance issue. I tried but failed to find a satisfying third-party (command-line driven) tool that can perform restraint optimization of the sugar-phosphate backbone geometry while keeping base atoms fixed.
The problem was finally solved after I attended the 43rd Mid-Atlantic Macromolecular Crystallography Meeting held at Duke University a few months ago. At the meeting, I had the opportunities to talk to several members of the PHENIX team. Particularly, Jeff Headd revised the geometry_minimization component of PHENIX to do the trick. Here is the mail reply from Jeff, using a 3DNA-generated DNA duplex (355d-3dna.pdb) as an example (see full details below):
Here’s a first go at refining just the backbone atoms of you input DNA model. You’ll need the most recently nightly build of Phenix (dev-1395 would work) and then run:
phenix.geometry_minimization 355d-3dna.pdb min.params
using the attached min.params file.
What I specify in the params file is to only move the backbone atoms, which I’ve done with a selection. You can modify the atoms that are allowed to move to your liking.
The only other change was to allow longer distance linkages, as some of the backbone linkages start quite far apart.
The content of file min.params is:
pdb_interpretation {
link_distance_cutoff = 7.0
}
selection = name " P " or name " OP1" or name " OP2" or \
name " O5'" or name " C5'" or name " C4'" or \
name " O4'" or name " C3'" or name " O3'" or \
name " C2'"
To make the story complete, given below is the step-by-step procedure, using 355d, a B-DNA dodecamer at 1.4 Å resolution as an example. The corresponding PDB file is named 355d.pdb.
find_pair 355d.pdb stdout | analyze stdin
x3dna_utils cp_std bdna
rebuild -atomic bp_step.par 355d-3dna.pdb
# the rebuilt structure is called '355d-3dna.pdb'
# with Phenix dev-1395 and above
phenix.geometry_minimization 355d-3dna.pdb min.params
# the optimized structure is called '355d-3dna_minimized.pdb'
# to verify:
find_pair 355d-3dna.pdb stdout | analyze stdin
find_pair 355d-3dna_minimized.pdb stdout | analyze stdin
# check files '355d-3dna.out' and '355d-3dna_minimized.out'
The three key files mentioned above are provided here for your verification:
Finally, the following figure illustrates the B-DNA dodecamer duplex in experimental (left), 3DNA-generated (middle) and PHENIX-optimized (right) coordinates. Note that disconnected O3′—P linkages (marked by red dots for two cases, see bottom of the middle image) due to overlong distances in 3DNA-rebuilt structure are fixed following the restraint PHENIX optimization.
| 355d-experimental |
3DNA-rebuilt |
PHENIX-optimized |
 |
 |
 |
Note added on 2016-11-11: In the min.params file, the selection is in one long line. For illustration purpose, the selection section (see below) is split into serveral short lines in the blog post. However, PHENIX requires ending backslashes (\) to combine the split lines into a single grammatical unit. I was not aware of this strict rule, and missed to add the ending \s in the original post. Thanks to Oleg Sobolev from the PHENIX team for pointing out this omission to my attention. Note that the content of min.params did not have a problem, and thus no change is made.
pdb_interpretation {
link_distance_cutoff = 7.0
}
selection = name " P " or name " OP1" or name " OP2" or \
name " O5'" or name " C5'" or name " C4'" or \
name " O4'" or name " C3'" or name " O3'" or \
name " C2'"

One of DSSR’s noteworthy features is the auto-detection of helical junctions in nucleic acids structures, be it RNA, DNA, or chimeric DNA/RNA, consisting of one or multiple chains. Helical junctions are created at the interface of three and more stems composed of canonical pairs (Watson-Crick A—T/U and G—C, or wobble G—U). A three-way junction model is illustrated below (copied from Figure 1 of the Bindewald et al. RNAJunction paper). Note that the three chains are each continuous (i.e., consecutive nts are covalently connected), and together with the three inner bps, forming a loop in the middle. Here, the three-way junction is of type [3×2×3], and the loop is composed of a total of 3×2+3+2+3 = 14 nts.

DSSR automatically detects all existing helical junctions in a nucleic acid structure, as illustrated by the following examples.
1l6b [all DNA Holliday junction structure of d(CCGGTACm5CGG)]
This is a simple four-way junction of type [0×0×0×0], where all bases are paired, leaving no connecting nts. The related portion of DSSR output is:
List of 1 junction(s)
1 4-way junctions: 8 nts; [0x0x0x0]; linked by [#1, #2, #4, #3]
1:A.DA6+1:A.DC7+2:B.DG14+2:B.DT15+2:A.DA6+2:A.DC7+1:B.DG14+1:B.DT15 [ACGTACGT]
0 nts junction ; 1:A.DA6-->1:A.DC7 [AC]
0 nts junction ; 2:B.DG14-->2:B.DT15 [GT]
0 nts junction ; 2:A.DA6-->2:A.DC7 [AC]
0 nts junction ; 1:B.DG14-->1:B.DT15 [GT]
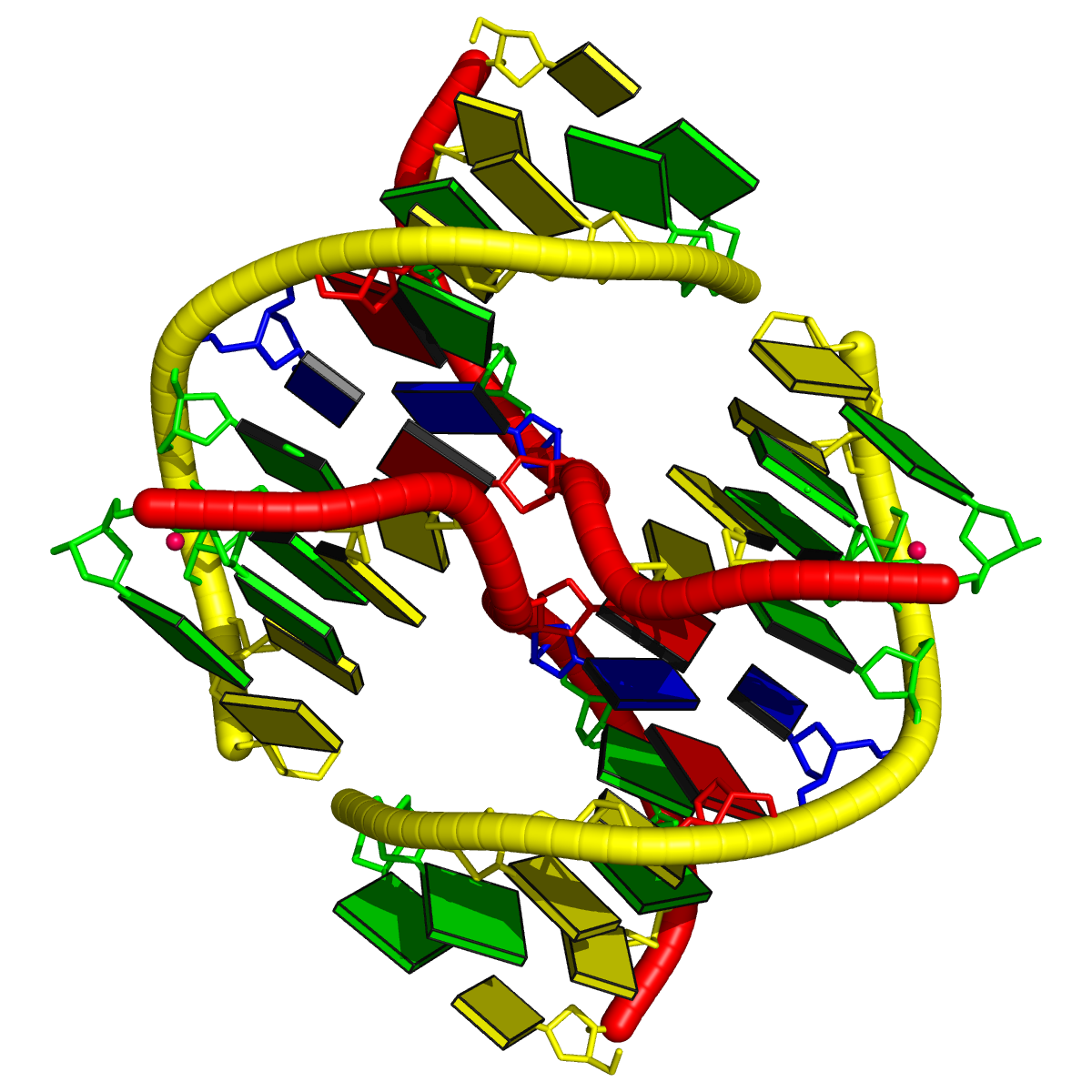
Technically, note the following points:
- The four-way junction is derived from the biological assembly 1 (PDB file
1l6b.pdb1), which contains two copies of the asymmetric unit, delineated by MODEL/ENDMDL. By default, DSSR/3DNA works one structure at a time, corresponding to the first structure/model in a given PDB or mmCIF file. To take the biological assembly as a whole, and to avoid confusions with MODEL/ENDMDL delineated NMR entries, the ENDMDL record of the first model is commented out in the file (1l6b.pdb1), as below:
#ENDMDL
MODEL 2
- With the modified PDB file
1l6b.pdb1, the DSSR command can be run as x3dna-dssr -i=1l6b.pdb1, with the output going to stdout.
- The simplified schematic block png image was generated with the command below to create the Raster3D
.r3d file (1l6b.r3d), which was then ray-traced using PyMOL.
blocview -r 1l6b.r3d 1l6b.pdb1
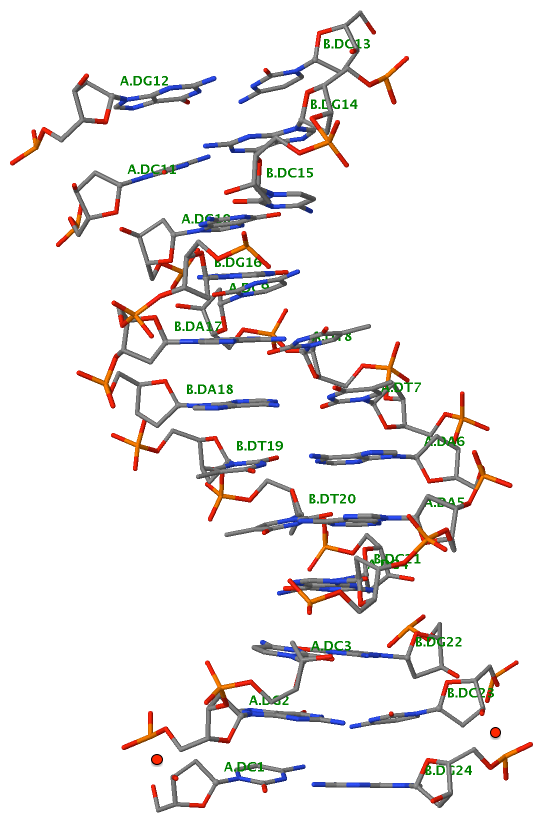
1egk [a four-way DNA/RNA junction]
This four-way junction consists of both DNA and RNA chains. Here the helical junction may not be that obvious by directly looking at the 3D image.
List of 1 junction(s)
1 4-way junctions: 10 nts; [0x0x1x1]; linked by [#3, #-1, #4, #5]
B.DC37+B.DT38+B.DA45+B.DC46+C.G109+C.A110+C.U111+D.DA130+D.DG131+D.DG132 [CTACGAUAGG]
0 nts junction ; B.DC37-->B.DT38 [CT]
0 nts junction ; B.DA45-->B.DC46 [AC]
1 nts junction C.A110 [A]; C.G109-->C.U111 [GAU]
1 nts junction D.DG131 [G]; D.DA130-->D.DG132 [AGG]
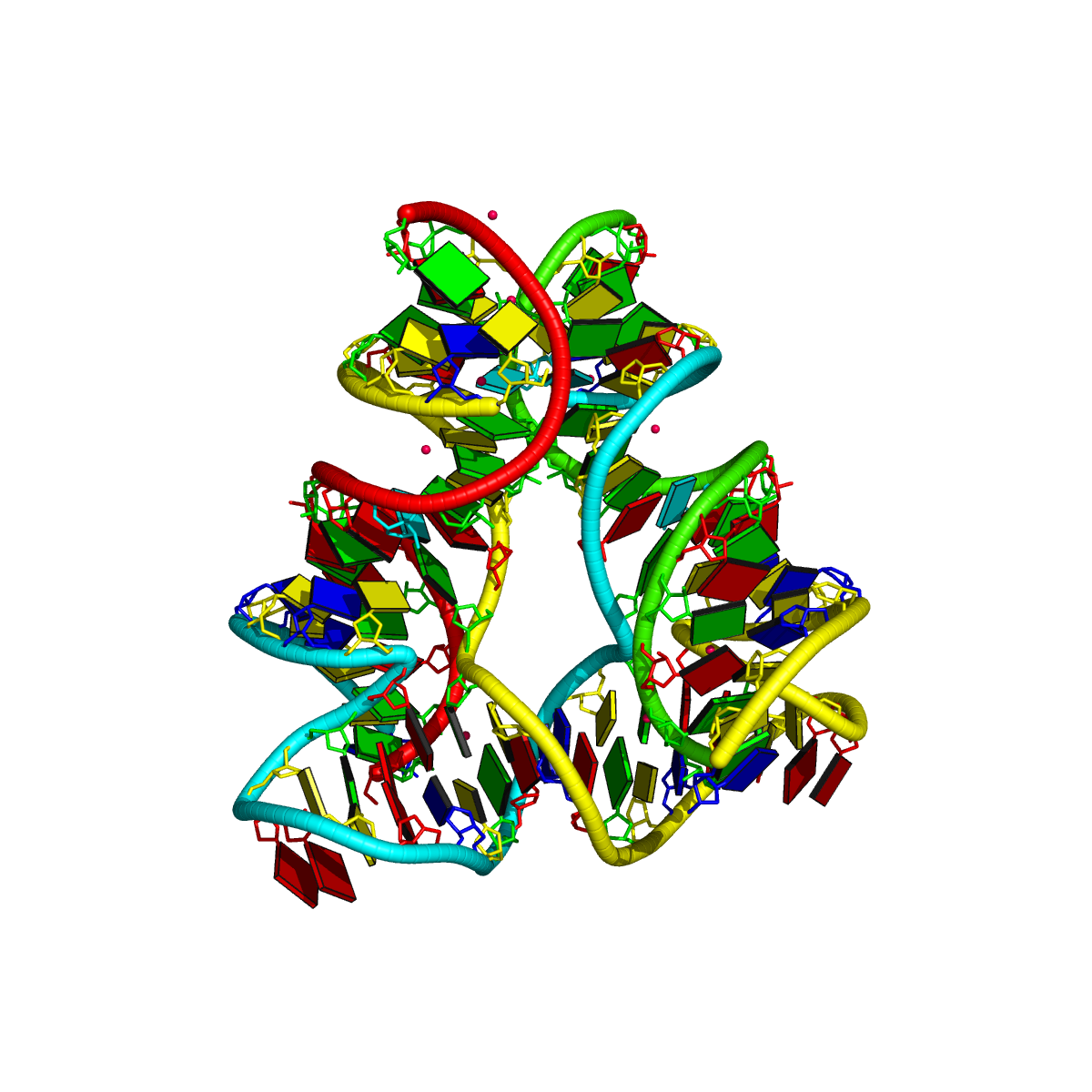
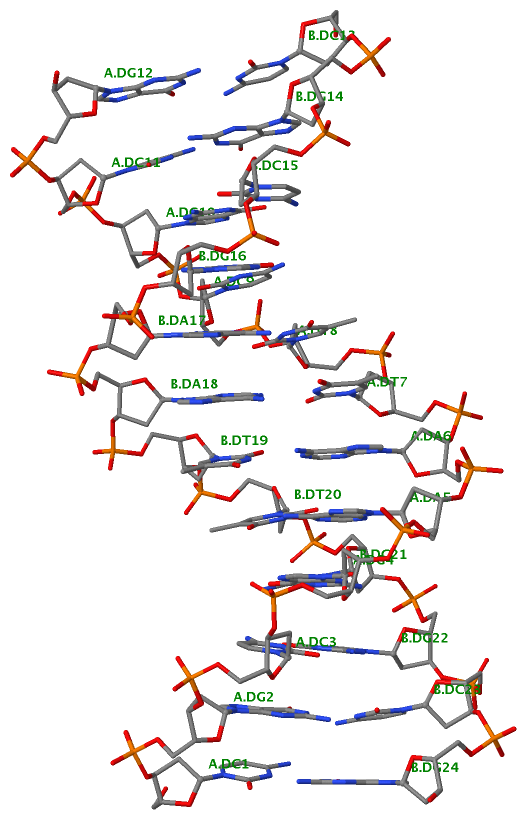
1ehz [yeast phenylalanine tRNA]
As shown below, DSSR correctly detects the classic L-shaped 3D structure and the cloverleaf 2D structure of a tRNA.
List of 1 junction(s)
1 4-way junctions: 16 nts; [2x1x5x0]; linked by [#1, #2, #3, #4]
A.U7+A.U8+A.A9+A.2MG10+A.C25+A.M2G26+A.C27+A.G43+A.A44+A.G45+A.7MG46+A.U47+A.C48+A.5MC49+A.G65+A.A66 [UUAgCgCGAGgUCcGA]
2 nts junction A.U8+A.A9 [UA]; A.U7-->A.2MG10 [UUAg]
1 nts junction A.M2G26 [g]; A.C25-->A.C27 [CgC]
5 nts junction A.A44+A.G45+A.7MG46+A.U47+A.C48 [AGgUC]; A.G43-->A.5MC49 [GAGgUCc]
0 nts junction ; A.G65-->A.A66 [GA]
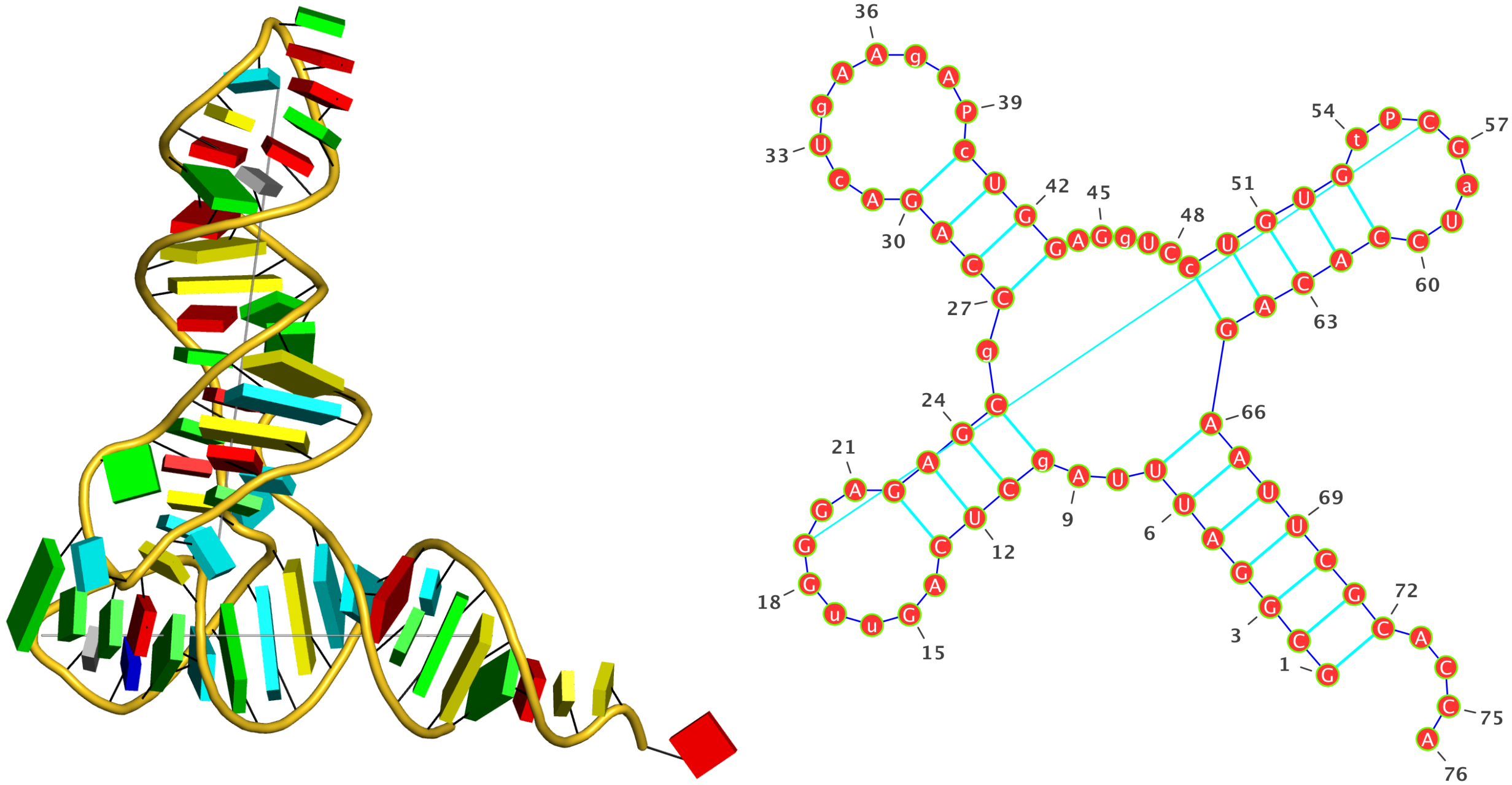
2fk6 [RNAse Z/tRNA(Thr) complex]
In a recent paper Predicting Helical Topologies in RNA Junctions as Tree Graphs by Laing et al., this PDB entry was selected in Table 1 as containing a three-way junction. However, DSSR fails to detect any junction in this structure, even though the program does find co-axial stacks. It turns out that the PDB entry 2fk6 does not possess the anti-codon stem/loop, thus nts C25 and G46 are not covalently connected. While three-way junctions may be defined differently, the DSSR result follows the above mentioned chain-continuity requirement.
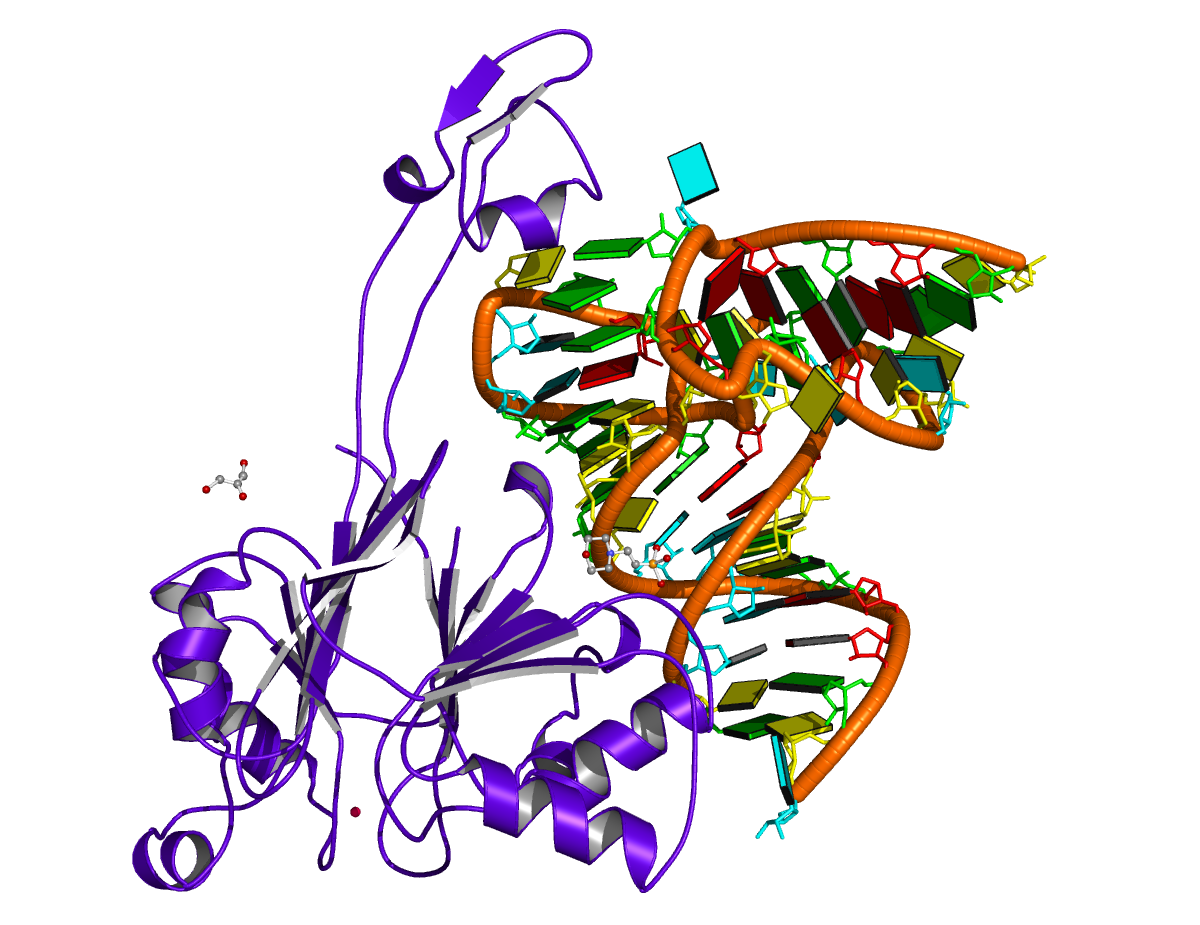
Overall, DSSR can consistently find all helical junctions in a given nucleic acid structure. Try DSSR on a ribosomal structure, you may well appreciate what it reveals. Moreover, it is straightforward to apply the program to all RNA/DNA-containing entries in the PDB via a script.

Given the primary sequence of an RNA molecule, there are numerous methods for predicting its secondary (2D) structures. To judge their accuracy, three-dimensional (3D) RNA structures solved experimentally by X-ray or NMR as deposited in the PDB are often used as benchmarks. DSSR is a handy tool to derive an RNA 2D structure from its 3D coordinates in PDB or mmCIF format. The 2D structure is specified in the dot-bracket notation (dbn), which can be fed directly into drawing programs such as VARNA for interactive display and easy generation of publication quality 2D diagrams.
Over the past few months, I’ve been asked a few times on the details of how the diagrams in the DSSR post were created. The answer is really simple, and has already been mentioned above and in the post. Here are two concrete examples to show how the process works.
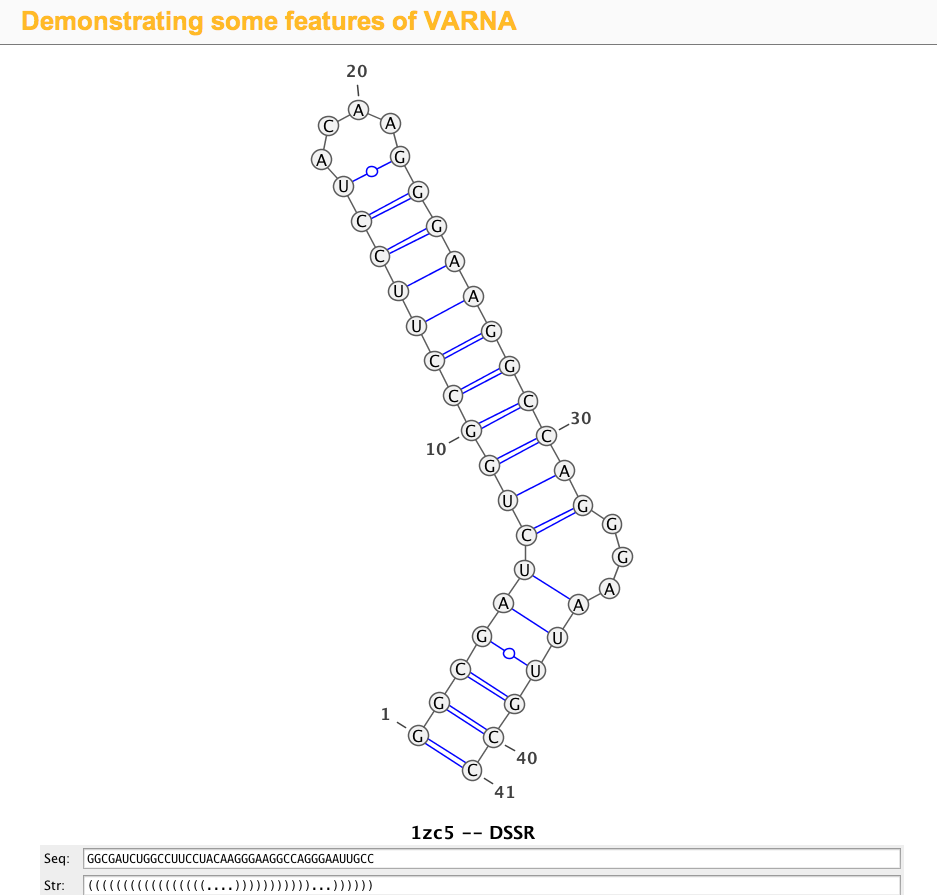
1zc5 (structure of the RNA signal essential for translational frame shifting in HIV-1)
This is the structure used in the VARNA paper. Let the PDB file be named 1zc5.pdb, the DSSR program can be run like this:
x3dna-dssr -i=1zc5.pdb
The output is sent to stdout by default, with the following three lines towards the end:
>1zc5-A #1 RNA with 41 nts
GGCGAUCUGGCCUUCCUACAAGGGAAGGCCAGGGAAUUGCC
(((((((((((((((((....)))))))))))...))))))
Simply copy and paste the last two lines (sequence and the 2D structure in dbn notation) into the Seq: and Str: fields of the VARNA demo page, the diagram will be updated automatically, as shown in the screenshot:
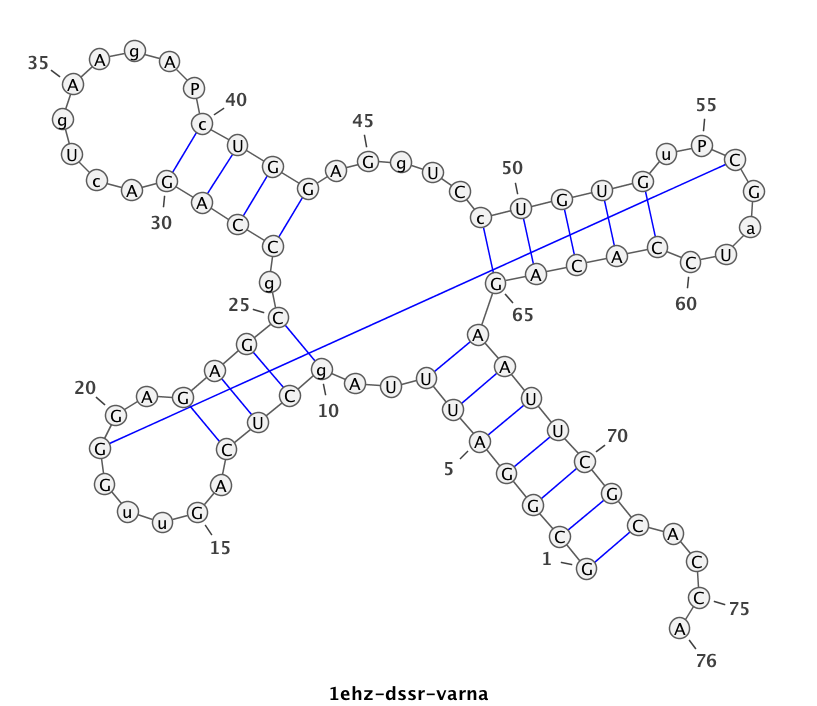
1ehz (crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution)
This example (1ehz.pdb) is used to illustrate tRNA’s classic cloverleaf 2D structure. The related command and result are:
x3dna-dssr -i=1ehz.pdb -o=1ehz.out
# the output is sent to file '1ehz.out'
# towards its end are the following 3 lines
>1ehz-A #1 RNA with 76 nts
GCGGAUUUAgCUCAGuuGGGAGAGCgCCAGAcUgAAgAPcUGGAGgUCcUGUGuPCGaUCCACAGAAUUCGCACCA
(((((((..((((.....[..)))).((((.........)))).....(((((..]....))))))))))))....
I’ve used a local copy of the JAVA web start version of VARNA (VARNA-WebStart.jnlp) to generate the following 2D diagram. Here, in addition to the customized title, I have set the number period to 5 nts, adopted the simple base-pair style, and manually adjusted the T arm (upper right corner) to make the long line connecting G19 and C56 a bit more unobtrusive. Right-click to see the context menu.
Note that the G19—C56 pair creates a pseudo-knot (specified by the matching [] pair in the dbn notation above) in tRNA. I was not aware of this salient feature from previous knowledge of relevant literature. It was indeed a surprise when I first saw it in the 2D diagram.
As illustrated above, DSSR serves well as a bridge from RNA 3D to 2D structures. Give DSSR a try, you will find the program actually has much more to offer!