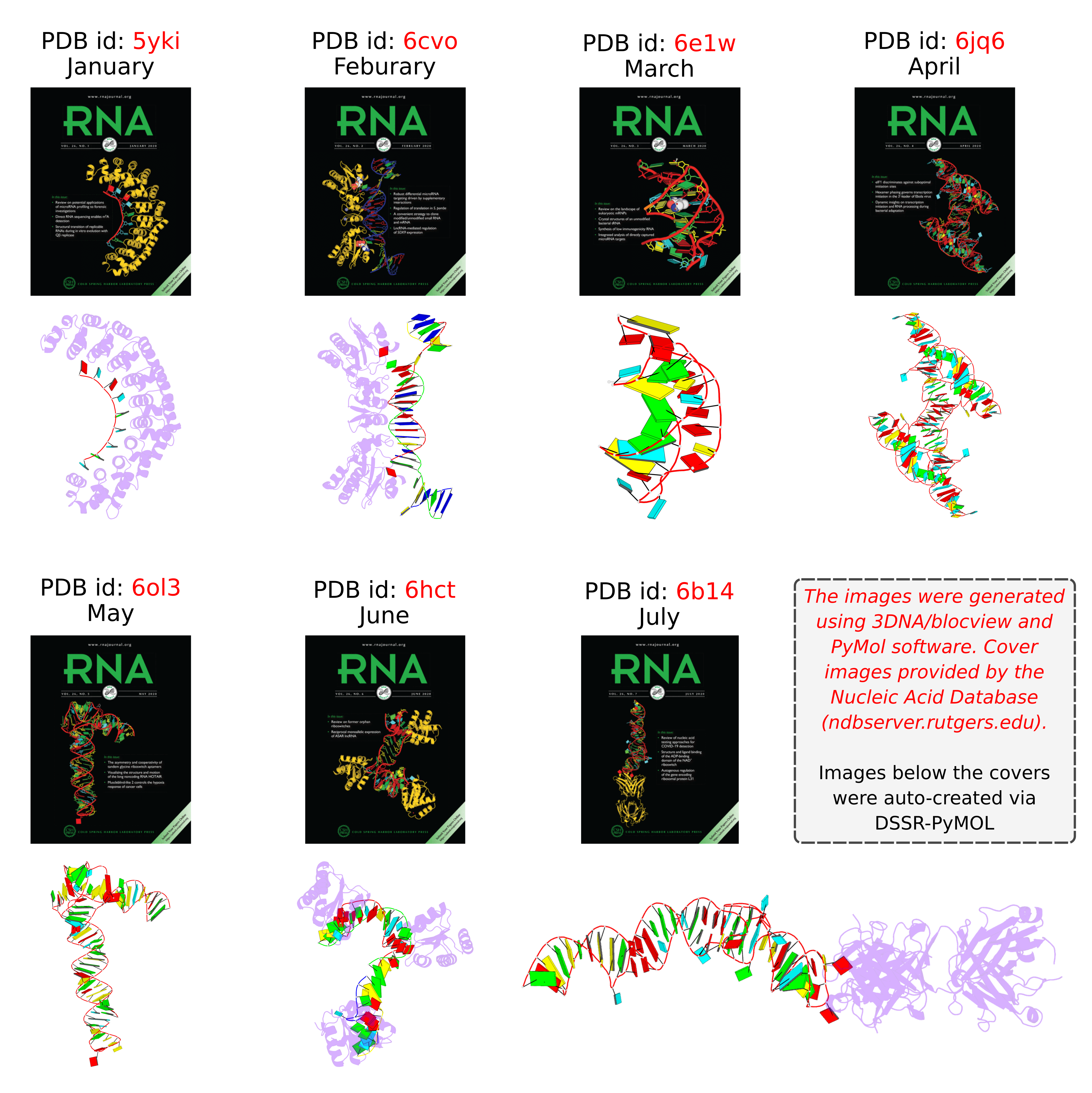
Following my previous post 3DNA/blocview-PyMOL images in covers of the RNA journal in 2019, here is an update for 2020. The cover images of the January to July issues have all been generated with help of 3DNA and provided by the NDB:
RNA is displayed as a red ribbon; block bases use NDB colors: A—red, C—yellow, G—green, U—cyan. The image was generated using 3DNA/blocview and PyMol software. Cover image provided by the Nucleic Acid Database (ndbserver.rutgers.edu).
Here is the composite figure of the seven cover images, with the brand new DSSR-PyMOL schematics for comparison.
Details of the seven structures illustrated in the cover images are described below:
- January 2020 Pumilio homolog PUF domain in complex with RNA (PDB id: 5yki; Zhao YY, Mao MW, Zhang WJ, Wang J, Li HT, Yang Y, Wang Z, Wu JW. 2018. Expanding RNA binding specificity and affinity of engineered PUF domains. Nucleic Acids Res 46: 4771–4782). Engineered nine-repeat PUF domain binds to its RNA target specifically and with high binding affinity.
- February 2020 Aprataxin RNA–DNA deadenylase product complex (PDB id: 6cvo; Tumbale P, Schellenberg MJ, Mueller GA, Fairweather E, Watson M, Little JN, Krahn J, Waddell I, London RE, Williams RS. 2018. Mechanism of APTX nicked DNA sensing and pleiotropic inactivation in neurodegenerative disease. EMBO J 37: e98875). Human aprataxin RNA–DNA deadenylase protects genome integrity and corrects abortive DNA ligation arising during ribonucleotide excision repair and base excision DNA repair.
- March 2020 PreQ1 riboswitch (PDB id: 6e1w; Connelly CM, Numata T, Boer RE, Moon MH, Sinniah RS, Barchi JJ, Ferre-D’Amare AR, Schneekloth Jr JS. 2019. Synthetic ligands for PreQ1 riboswitches provide structural and mechanistic insights into targeting RNA tertiary structure. Nat Commun 10: 1501). Class I PreQ1 riboswitch regulates downstream gene expression in response to its cognate ligand PreQ1 (7-aminomethyl-7-deazaguanine).
- April 2020 Hatchet ribozyme (PDB id: 6jq6; Zheng L, Falschlunger C, Huang K, Mairhofer E, Yuan S, Wang J, Patel DJ, Micura R, Ren A. 2019. Hatchet ribozyme structure and implications for cleavage mechanism. Proc Natl Acad Sci 116: 10783–10791). This crystal structure of the hatchet ribozyme product features a compact symmetric dimer.
- May 2020 Adenovirus virus-associated RNA (PDB id: 6ol3; Hood IV, Gordon JM, Bou-Nader C, Henderson FE, Bahmanjah S, Zhang J. 2019. Crystal structure of an adenovirus virus-associated RNA. Nat Commun 10: 2871). Acutely bent viral RNA fragment is a protein kinase R inhibitor and features an unusually structured apical loop, a wobble-enriched, coaxially stacked apical and tetra-stems, and a central domain pseudoknot that resembles codon-anticodon interactions.
- June 2020 Archeoglobus fulgidus L7Ae bound to cognate K-turn (PDB id: 6hct; Huang L, Ashraf S, Lilley DMJ. 2019. The role of RNA structure in translational regulation by L7Ae protein in archaea. RNA 25: 60–69). 50S archaeal ribosome protein L7Ae binds to a K-turn structure in the 5′-leader of the mRNA of its structural gene to regulate translation.
- July 2020 Spinach RNA aptamer/Fab complex (PDB id: 6b14; Koirala D, Shelke SA, Dupont M, Ruiz S, DasGupta S, Bailey LJ, Benner SA, Piccirilli JA. 2018. Affinity maturation of a portable Fab-RNA module for chaperone-assisted RNA crystallography. Nucleic Acids Res 46: 2624–2635). Novel Fab-RNA module can serve as an affinity tag for RNA purification and imaging and as a chaperone for RNA crystallography.