Dot bracket notation (dbn) is a popular format to represent RNA secondary structures. Initially introduced by the ViennaRNA package, dbn uses dots (.) for unpaired bases, and matched parentheses () for the canonical Watson-Crick A-T and G-C or the wobble G-U pairs. This compact representation was designed for fully nested (i.e., pseudoknot free) RNA secondary structures in a single RNA molecule. Over the years, it has been extended to cover pseudoknots (of possibly higher orders) using matched pairs of [], {}, and <> etc.
To derive dbn from three-dimensional atomic coordinates with DSSR, I was faced with an issue on how to represent multiple RNA chains (molecules). A closely related yet practical problem is chain breaks, as in x-ray crystal structures where disordered regions may not have fitted coordinates. I searched but failed to find any ‘standard’ way to account for chain breaks or multiple molecules in dbn. The commonly used programs for visualizing RNA secondary structure diagrams that I tested at that time did not take such cases into consideration — they simply showed all bases as if they were from a single continous RNA chain.
I discussed the issue with Dr. Yann Ponty, the maintainer of the popular VARNA program. After a few around of email exchanges, we introduced an extra symbol (&) in both sequence and dbn to designate multiple chains or breaks within a chain to communicate between DSSR and VARNA.
As an example, the DSSR-derived dbn for the double-stranded DNA structure 355d (the famous Dickerson dodecamer) is as below:
Secondary structures in dot-bracket notation (dbn) as a whole and per chain >355d nts=24 [whole] CGCGAATTCGCG&CGCGAATTCGCG ((((((((((((&)))))))))))) >355d-A #1 nts=12 [chain] DNA CGCGAATTCGCG (((((((((((( >355d-B #2 nts=12 [chain] DNA CGCGAATTCGCG ))))))))))))
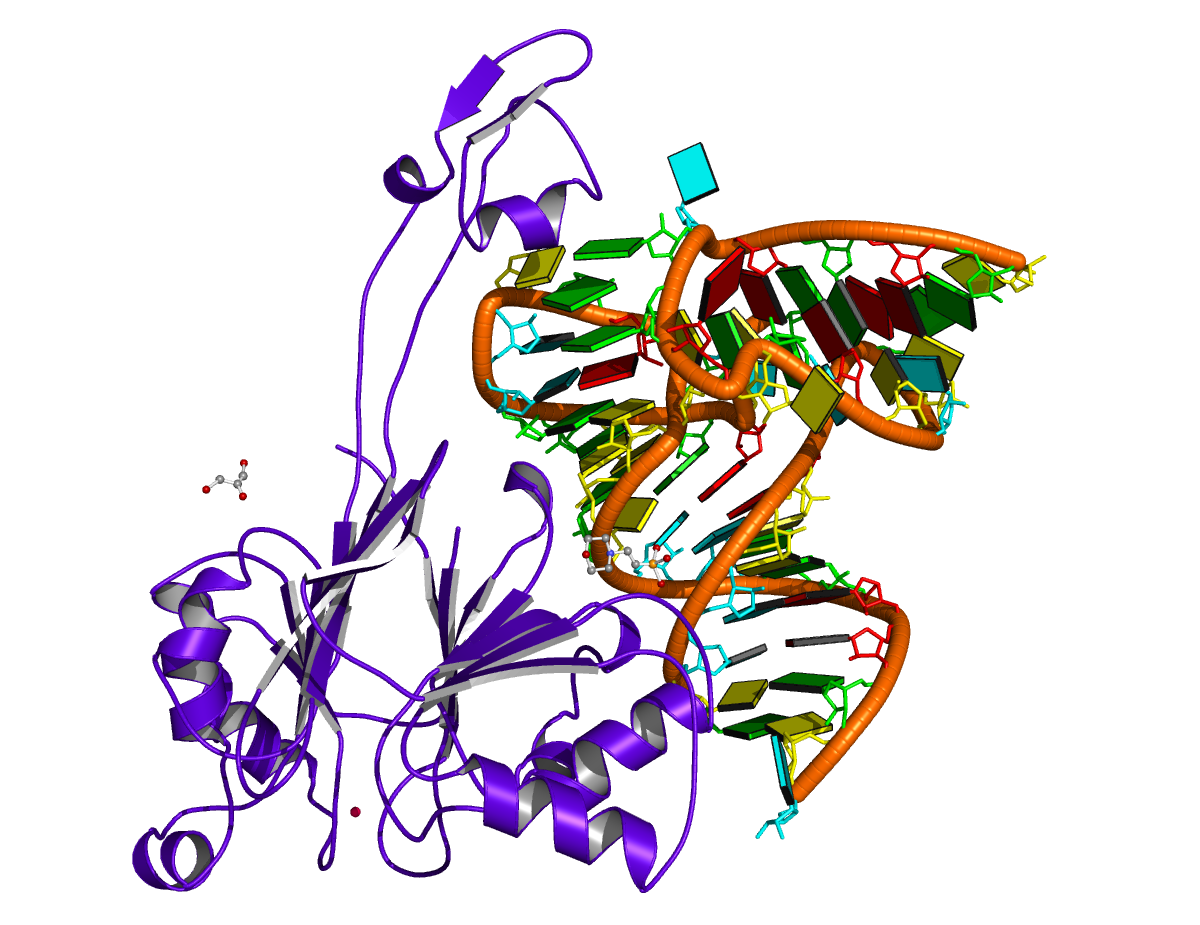
As another example, the PDB entry 2fk6 contains a tRNA with chain breaks — nucleotides 26 to 45 are missing from the structure (see figure below). The DSSR-derived dbn is as follows — note the * at the end of the header line.
>2fk6-R #1 nts=53 [chain] RNA* GCUUCCAUAGCUCAGCAGGUAGAGC&GUCAGCGGUUCGAGCCCGCUUGGAAGCU (((((((..((((.....[..))))&...(((((..]....)))))))))))).
It is worth mentioning a subtle point in DSSR-derived dbn with multiple chains, i.e., the order of the chains may make a difference! The point is best illustrated with a concrete example — here, 4un3, the crystal structure of Cas9 bound to PAM-containing DNA target. Based on the data file downloaded directly from the PDB (4un3.pdb), the relevant portions of DSSR output are:
****************************************************************************
Special notes:
o Cross-paired segments in separate chains, be *careful* with .dbn
****************************************************************************
This structure contains *1-order pseudoknot
o You may want to run DSSR again with the '--nested' option which removes
pseudoknots to get a fully nested secondary structure representation.
o The DSSR-derived dbn may be problematic (see notes above).
****************************************************************************
Secondary structures in dot-bracket notation (dbn) as a whole and per chain
>4un3 nts=120 [whole]
AUAACUCAAUUUGUAAAAAAGUUUUAGAGCUAGAAAUAGCAAGUUAAAAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUG&CAATACCATTTTTTACAAATTGAGTTAT&AAATGGTATTG
((((((((((((((((((((((((((..((((....))))....))))))..(((..).)).......((((....)))).&[[[[[[[[))))))))))))))))))))&...]]]]]]]]
>4un3-A #1 nts=81 [chain] RNA
AUAACUCAAUUUGUAAAAAAGUUUUAGAGCUAGAAAUAGCAAGUUAAAAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUG
((((((((((((((((((((((((((..((((....))))....))))))..(((..).)).......((((....)))).
>4un3-C #2 nts=28 [chain] DNA
CAATACCATTTTTTACAAATTGAGTTAT
[[[[[[[[))))))))))))))))))))
>4un3-D #3 nts=11 [chain] DNA
AAATGGTATTG
...]]]]]]]]
The notes in the DSSR output is worth paying attention to. Specifically, it reports a “*1-order pseudoknot” — note also the *! Here the target DNA chain C comes before DNA chain D in the PDB file. The 5′-end bases in chain C pair with bases in D, and the 3′-end bases in C pair with RNA bases in chain A. There exist pairs crossing along the ‘linear’ sequence position-wise, hence the reported “pseudoknot”. However, simply reverse DNA chains C and D, i.e., moving chain D before C (in file 4un3-ADC.pdb), the “pseudoknot” will be gone, as shown below:
**************************************************************************** Secondary structures in dot-bracket notation (dbn) as a whole and per chain >4un3-ADC nts=120 [whole] AUAACUCAAUUUGUAAAAAAGUUUUAGAGCUAGAAAUAGCAAGUUAAAAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUG&AAATGGTATTG&CAATACCATTTTTTACAAATTGAGTTAT ((((((((((((((((((((((((((..((((....))))....))))))..(((..).)).......((((....)))).&...((((((((&)))))))))))))))))))))))))))) >4un3-ADC-A #1 nts=81 [chain] RNA AUAACUCAAUUUGUAAAAAAGUUUUAGAGCUAGAAAUAGCAAGUUAAAAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUG ((((((((((((((((((((((((((..((((....))))....))))))..(((..).)).......((((....)))). >4un3-ADC-D #2 nts=11 [chain] DNA AAATGGTATTG ...(((((((( >4un3-ADC-C #3 nts=28 [chain] DNA CAATACCATTTTTTACAAATTGAGTTAT ))))))))))))))))))))))))))))
Notes added on March 19, 2015
- It has drawn to my attention that the NUPACK uses ‘+’ instead of ‘&’ as the symbol to separate multiple chains (or chain breaks). In fact, DSSR has an undocumented option
--dbn_breakwhich can be set to any of the character in the string&.:,|+. The ‘&’ symbol was chosen for communication with VARNA which requires ‘&’, at least up to now. This is an excellent example showing the efforts that I have put into the little details while developing DSSR.
- The issue on proper ordering of multiple chains to avoid crossing lines (false pseudoknots) has been formally addressed by Dirks et al. in their 2007 article titled Thermodynamic analysis of interacting nucleic acid strands (SIAM Rev, 49, 65-88), specifically in Section 2.1 (Fig. 2.1). Applying that algorithm to nucleic acid structures, however, is beyond the scope of DSSR. The program strictly respects the ordering of chains and nucleotides within a given PDB or PDBx/mmCIF file, but outputs warning messages where necessary to draw users’ attention. As another example, I’ve recently noticed that DNA duplexes produced by Maestro (a product of Schrödinger) list nucleotides of the complementary strand in 3′ to 5′ order to match the 5′ to 3′ directionality of the leading strand for each Watson-Crick pair (See below).
**************************************************************************** Special notes: o nucleotides out of order **************************************************************************** Secondary structures in dot-bracket notation (dbn) as a whole and per chain >ga62_ca62_1m_in nts=24 [whole] GGCGAATTCCGG&C&C&G&C&T&T&A&A&G&G&C&C ((((((((((((&)&)&)&)&)&)&)&)&)&)&)&) >ga62_ca62_1m_in-1-A #1 nts=12 [chain] DNA GGCGAATTCCGG (((((((((((( >ga62_ca62_1m_in-1-B #2 nts=12 [chain] DNA C&C&G&C&T&T&A&A&G&G&C&C )&)&)&)&)&)&)&)&)&)&)&)