Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. [Nucleic Acids Res 48: e74(https://doi.org/10.1093/nar/gkaa426)).
See the 2020 paper titled "DSSR-enabled innovative schematics of 3D nucleic acid structures with PyMOL" in Nucleic Acids Research and the corresponding Supplemental PDF for details. Many thanks to Drs. Wilma Olson and Cathy Lawson for their help in the preparation of the illustrations.
Details on how to reproduce the cover images are available on the 3DNA Forum.

Cryo-EM structure of the pre-B complex (PDB id: 8QP8; Zhang Z, Kumar V, Dybkov O, Will CL, Zhong J, Ludwig SE, Urlaub H, Kastner B, Stark H, Lührmann R. 2024. Structural insights into the cross-exon to cross-intron spliceosome switch. Nature 630: 1012–1019). The pre-B complex is thought to be critical in the regulation of splicing reactions. Its structure suggests how the cross-exon and cross-intron spliceosome assembly pathways converge. The U4, U5, and U6 snRNA backbones are depicted respectively by blue, green, and red ribbons, with bases and Watson-Crick base pairs shown as color-coded blocks: A/A-U in red, C/C-G in yellow, G/G-C in green, U/U-A in cyan; the proteins are represented by gold ribbons. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Structure of the Hendra henipavirus (HeV) nucleoprotein (N) protein-RNA double-ring assembly (PDB id: 8C4H; Passchier TC, White JB, Maskell DP, Byrne MJ, Ranson NA, Edwards TA, Barr JN. 2024. The cryoEM structure of the Hendra henipavirus nucleoprotein reveals insights into paramyxoviral nucleocapsid architectures. Sci Rep 14: 14099). The HeV N protein adopts a bi-lobed fold, where the N- and C-terminal globular domains are bisected by an RNA binding cleft. Neighboring N proteins assemble laterally and completely encapsidate the viral genomic and antigenomic RNAs. The two RNAs are depicted by green and red ribbons. The U bases of the poly(U) model are shown as cyan blocks. Proteins are represented as semitransparent gold ribbons. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).

Structure of the helicase and C-terminal domains of Dicer-related helicase-1 (DRH-1) bound to dsRNA (PDB id: 8T5S; Consalvo CD, Aderounmu AM, Donelick HM, Aruscavage PJ, Eckert DM, Shen PS, Bass BL. 2024. Caenorhabditis elegans Dicer acts with the RIG-I-like helicase DRH-1 and RDE-4 to cleave dsRNA. eLife 13: RP93979. Cryo-EM structures of Dicer-1 in complex with DRH-1, RNAi deficient-4 (RDE-4), and dsRNA provide mechanistic insights into how these three proteins cooperate in antiviral defense. The dsRNA backbone is depicted by green and red ribbons. The U-A pairs of the poly(A)·poly(U) model are shown as long rectangular cyan blocks, with minor-groove edges colored white. The ADP ligand is represented by a red block and the protein by a gold ribbon. Cover image provided by X3DNA-DSSR, an NIGMS National Resource for structural bioinformatics of nucleic acids (R24GM153869; skmatics.x3dna.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).
Moreover, the following 30 [12(2021) + 12(2022) + 6(2023)] cover images of the RNA Journal were generated by the NAKB (nakb.org).
Cover image provided by the Nucleic Acid Database (NDB)/Nucleic Acid Knowledgebase (NAKB; nakb.org). Image generated using DSSR and PyMOL (Lu XJ. 2020. Nucleic Acids Res 48: e74).
It gives me great pleasure to announce that the 3DNA/DSSR project is now funded by the NIH R24GM153869 grant, titled "X3DNA-DSSR: a resource for structural bioinformatics of nucleic acids". I am deeply grateful for the opportunity to continue working on a project that has basically defined who I am. It was a tough time during the funding gap over the past few years. Nevertheless, I have experienced and learned a lot, and witnessed miracles enabled by enthusiastic users.
Since late 2020 when I lost my R01 grant, DSSR has been licensed by the Columbia Technology Ventures (CTV). I appreciate the numerous users (including big pharma) who purchased a DSSR Pro License or a DSSR Basic paid License. Thanks to the NIH R24GM153869 grant, we are pleased to provide DSSR Basic free of charge to the academic community. Academic Users may submit a license request for DSSR Basic or DSSR Pro by clicking "Express Licensing" on the CTV landing page. Commercial users may inquire about pricing and licensing terms by emailing techtransfer@columbia.edu, copying xiangjun@x3dna.org.
DSSR v2.4.5-2024sep24 was released to synchronize with the new R24 funding, which will bring the project to an entirely new level. All existing users are encouraged to upgrade their installation to this release which contains miscellaneous bug fixes (e.g., chain id with > 4 chars) and numerous minor improvements.
Lots of exciting things will happen for the project. The first thing is to make DSSR freely accessible to the academic community. In the past couple of weeks, CTV have already issued quite a few DSSR Basic Academic licenses to users from all over the world. So the demand is high, and it will become stronger as more academic users become aware of DSSR. I'm closely monitoring the 3DNA Forum, and is always ready to answer users questions.
I am committed to making DSSR a brand that stands for quality and value. By virtue of its unmatched functionality, usability, and support, DSSR saves users a substantial amount of time and effort when compared to other options. My track record throughout the years has unambiguously demonstrated my dedication to this solid software product.
DSSR Basic contains all features described in the three DSSR-related papers, and includes the originally separate SNAP program (still unpublished) for analyzing DNA/RNA-protein complexes. The Pro version integrates the classic 3DNA functionality, plus advanced modeling routines, with email/Zoom/phone support.
Over the years, I have played quite a few computer programming languages. ANSI C has become my top choice for ‘serious’ software projects, due to its small size, efficiency, flexibility, and ubiquitous support. Moreover, C is a mature language, with a rich ecosystem. As it turns out, C has also been consistently rated as one of the most popular computer languages (#1 or #2) over the past thirty years.
Needless to say, ANSI C has its own quirks, and it takes a steep learning curve. However, once you get over the hurdles, the language serves you. I cannot remember when, but it has been a long while that coding in ANSI C is no longer an issue. It is the understanding of scientific questions that takes most of my time, and coding helps greatly in refining my thoughts.
Not surprisingly, ANSI C was chosen as the sole language for DSSR (and SNAP, or 3DNA in general). The ensure the overall quality of the DSSR codebase, I have taken the following steps:
- The whole project is under git.
- The ANSI C source code is compiled with strict GCC options for full compliance to the standard:
-ansi -pedantic -W -Wall -Wextra -Wunused -Wshadow -Werror -O3
- The executable is checked with valgrind for any memory leak:
valgrind --leak-check=full x3dna-dssr -i=1ehz.pdb -o=1ehz.out --quiet
==19624== Memcheck, a memory error detector
==19624== Copyright (C) 2002-2013, and GNU GPL'd, by Julian Seward et al.
==19624== Using Valgrind-3.10.1 and LibVEX; rerun with -h for copyright info
==19624== Command: x3dna-dssr -i=1ehz.pdb -o=1ehz.out --quiet
==19624==
==19624==
==19624== HEAP SUMMARY:
==19624== in use at exit: 0 bytes in 0 blocks
==19624== total heap usage: 52,829 allocs, 52,829 frees, 92,878,578 bytes allocated
==19624==
==19624== All heap blocks were freed -- no leaks are possible
==19624==
==19624== For counts of detected and suppressed errors, rerun with: -v
==19624== ERROR SUMMARY: 0 errors from 0 contexts (suppressed: 0 from 0)
- Extensive tests (with the simple
diff command) to ensure the program is working as expected.
The above four measures combined allow me to add new features, refactor the code, and fix bugs, without worrying about accidentally breaking existing functionality. Reading literature (including citations to 3DNA/DSSR) and responding to user feedback on the 3DNA Forum keep me continuously improve DSSR. Some of the recent refinements to DSSR came about this way.

Recently, I became aware of the metallo-base pairs, such as T-Hg-T (PDB id: 4l24) and C-Ag-C (5ay2) from the work of Kondo et al (Pubmed: 24478025 and 26448329). As of v1.4.3-2015oct23, DSSR can detect such metallo-bps automatically, as shown below:
# x3dna-dssr -i=4l24.pdb -o=4l24.out
List of 12 base pairs
nt1 nt2 bp name Saenger LW DSSR
1 A.DC1 B.DG24 C-G WC 19-XIX cWW cW-W
2 A.DG2 B.DC23 G-C WC 19-XIX cWW cW-W
3 A.DC3 B.DG22 C-G WC 19-XIX cWW cW-W
4 A.DG4 B.DC21 G-C WC 19-XIX cWW cW-W
5 A.DA5 B.DT20 A-T WC 20-XX cWW cW-W
6 A.DT6 B.DT19 T-T Metal n/a cWW cW-W
7 A.DT7 B.DT18 T-T Metal n/a cWW cW-W
8 A.DT8 B.DA17 T-A WC 20-XX cWW cW-W
9 A.DC9 B.DG16 C-G WC 19-XIX cWW cW-W
10 A.DG10 B.DC15 G-C WC 19-XIX cWW cW-W
11 A.DC11 B.DG14 C-G WC 19-XIX cWW cW-W
12 A.DG12 B.DC13 G-C WC 19-XIX cWW cW-W
and
# x3dna-dssr -i=5ay2.pdb -o=5ay2.out
List of 24 base pairs
nt1 nt2 bp name Saenger LW DSSR
1 A.G1 B.C12 G-C WC 19-XIX cWW cW-W
2 A.G2 B.C11 G-C WC 19-XIX cWW cW-W
3 A.A3 B.U10 A-U WC 20-XX cWW cW-W
4 A.C4 B.C9 C-C Metal n/a cWW cW-W
5 A.U5 B.A8 U-A WC 20-XX cWW cW-W
6 A.CBR6 B.G7 c-G WC 19-XIX cWW cW-W
7 A.G7 B.CBR6 G-c WC 19-XIX cWW cW-W
8 A.A8 B.U5 A-U WC 20-XX cWW cW-W
9 A.C9 B.C4 C-C Metal n/a cWW cW-W
10 A.U10 B.A3 U-A WC 20-XX cWW cW-W
11 A.C11 B.G2 C-G WC 19-XIX cWW cW-W
12 A.C12 B.G1 C-G WC 19-XIX cWW cW-W
13 C.G1 D.C12 G-C WC 19-XIX cWW cW-W
14 C.G2 D.C11 G-C WC 19-XIX cWW cW-W
15 C.A3 D.U10 A-U WC 20-XX cWW cW-W
16 C.C4 D.C9 C-C Metal n/a cWW cW-W
17 C.U5 D.A8 U-A WC 20-XX cWW cW-W
18 C.CBR6 D.G7 c-G WC 19-XIX cWW cW-W
19 C.G7 D.CBR6 G-c WC 19-XIX cWW cW-W
20 C.A8 D.U5 A-U WC 20-XX cWW cW-W
21 C.C9 D.C4 C-C Metal n/a cWW cW-W
22 C.U10 D.A3 U-A WC 20-XX cWW cW-W
23 C.C11 D.G2 C-G WC 19-XIX cWW cW-W
24 C.C12 D.G1 C-G WC 19-XIX cWW cW-W
Note the name “Metal” for the metallo-bps. Moreover, the corresponding entries in the ‘dssr-pairs.pdb’ file also include the metal ions, as shown below:
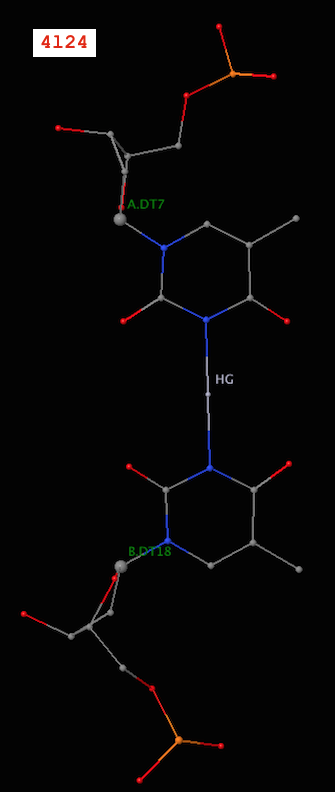
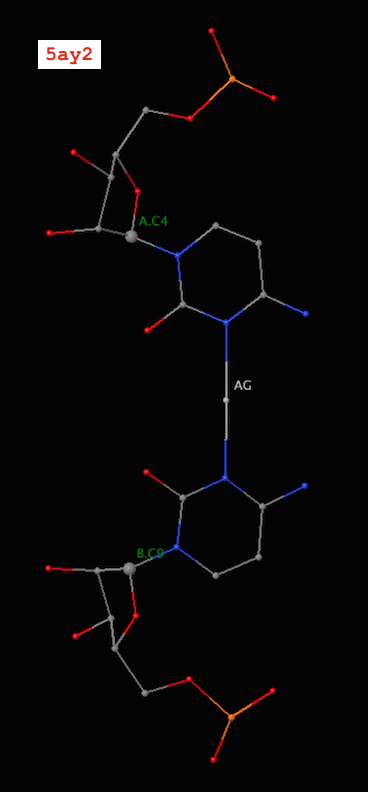
It is worth noting that in a metallo-bp, the metal ion lies approximately in the bp plane. Moreover, it is in the middle of the two bases, which would otherwise not form a pair in the conventional sense.

From the Jmol mailing list, I noticed Jmol 14.4.0 was released yesterday (October 13, 2015) by Dr. Bob Hanson. Among the development highlights is the following item:
biomolecule annotations including DSSR, RNA3D, EBI sequence domains, and PDB validation data
I am glad to see that DSSR has been integrated into Jmol, one of the most popular molecular graphics visualization programs. To enable easy access to the DSSR functionality from Jmol, I’ve set up two websites with easy-to-remember URLs: http://jmol.x3dna.org and http://jsmol.x3dna.org. They both point to the same jsmol/ folder extracted from jsmol.zip of the Jmol distribution.
In retrospect, I first met Bob at the Workshop on the PDBx/mmCIF Data Exchange Format for Structural Biology held at Rutgers University during October 21-22, 2013. I approached him during a lunch break, asking for a possible collaboration on integrating DSSR into Jmol. The name DSSR may have played a role in convincing Bob, since it matches the well-known DSSP program for proteins. In the end, we were both excited about the project, talked into details after the meeting, and continued our conversation the next morning while I drove him to the airport.
Nothing real happened until early April 2014. Once getting started, however, we moved forward rapidly: it took less then three weeks to get the first functional version ready for the community to play. See Bob’s announcement RNA/DNA Secondary Structure, anyone? in the Jmol mailing list on April 9, 2014. During this process, we communicated extensively via email, up to 30 messages per day, on technical details for better communication between the two programs. The integration works by using Jmol as a front-end, which calls a web-serivce hosted at Columbia University for DSSR analysis. Jmol’s parsing of the DSSR output is facilitated by the dedicated --jmol option.
The above preliminary, yet functional, DSSR-Jmol integration had be in service without infrastructural changes until two months ago. In August 10, 2015, Bob contacted me:
I might make a significant request though. That would be for the server to deliver all this in JSON format. This is really the way to go. It is what people want and it is perfect for Jmol as well.
I’d played around with JSON or SQLite as a structured data exchange format for quite some time, and Bob’s request finally convinced me that JSON is the (better) way to go. And that began another around of intensive collaborative work that has switched the exchange format between DSSR and Jmol from plain text output to JSON. From August 10 to September 22, we had a total of over 170-email exchanges, plus Skype. JSON has really simplified lives of both parties, especially in the long run.
Overall, collaborating with Bob has been truly an enjoyable and rewarding experience. The DSSR-Jmol integration also serves as a concrete example of what can be achieved by two dedicated minds with complementary expertise.

Curves+ and 3DNA are currently the most widely used programs for analyzing nucleic acid structures (predominantly double helices). As noted in my blog post, Curves+ vs 3DNA, these two programs also complement each other in terms of features. It thus makes sense to run both to get a better understanding of the DNA/RNA structures one is interested in.
Indeed, over the past few years, I have seen quite a few articles citing both 3DNA and Curves+. Listed below are three recent examples:
The helical parameters were measured with 3DNA33 and Curves+.34 The local helical parameters are defined with regard to base steps and without regard to a global axis.
Structure analysis. Helix, base and base pair parameters were calculated with 3DNA or curve+ software packages23,24.
The major global difference between the native and mixed backbone structures is that the RNA backbone is compressed or kinked in strands containing the modified linkage (Fig. 3 B and C, by CURVES) (30). … To compare the three RNA structures at a more detailed and local level, we calculated the base pair helical and step parameters for all three structures using the 3DNA software tools (31) (Fig. 4 and Table S2). [In the Results section]
For each snapshot, the structural parameters—including six base pair parameters, six local base pair step parameters, and pseudorotation angles for each nucleotide—were calculated using 3DNA (31). The two terminal base pairs are omitted for the 3DNA analysis, because they unwind frequently in the triple 2′-5′-linked duplex. [In the Materials and Methods section]
Reading through these papers, however, it is not clear to me if the authors took advantage of the find_pair -curves+ option in 3DNA, as detailed in Building a bridge between Curves+ and 3DNA. Hopefully, this post will help draw more attention to this connection between Curves+ and 3DNA.

Over the past couple of weeks, I’ve added two more DSSR options, --symmetry and --nmr, that are closely related to an ensemble of MODEL/ENDMDL-delineated structures in PDB files. However, there exist subtle differences between the two cases, and the usage of the same MODEL/ENDMDL ensemble format can be ambiguous to the uninitiated. This blog post aims to clarify the issues, using concrete examples.
The --symmetry options applies to X-ray crystal structures where an asymmetric unit represents only part of the whole biological assembly. In standard PDB format, the asymmetric unit contains instructions to produce crystallographic symmetry
related molecules.. Nevertheless, the biological assembly are also provided by the PDB (or NDB), with coordinate files ending with .pdb1 or such. For example, the PDB entry 2d94 has the single-stranded sequence GGGCGCCC in its asymmetric unit (2d94.pdb). It is the biological assembly in file 2d94.pdb1 that contains the DNA double helix.
x3dna-dssr -i=2d94.pdb # no pairs found
x3dna-dssr -i=2d94.pdb1 # still no pairs found
x3dna-dssr -i=2d94.pdb1 --symm # 8 pairs found
x3dna-dssr -i=2d94.pdb --symm # no pairs found
As shown by the above examples, DSSR by default reads only the first model even given the biological assemble file 2d94.pdb1. It is with --symmetry (abbreviated to --symm) explicitly specified that DSSR takes all models in the input biological assemble file into consideration. The last case also illustrates that DSSR does not generate crystallographic symmetry related molecules. The --symm simply informs DSSR to take all models, which already exist in the input file, into consideration.
On the other hand, the --nmr option is for auto-processing an ensemble of structures solved by solution NMR method (or trajectories of molecular dynamics simulations). The key point here is that each of the MODEL/ENDMDL-delinated structures is independent and thus can be processed separately, even though they are obviously closely related. Using the PDB entry 2n2d as an example, here are some sample usages:
x3dna-dssr -i=2n2d.pdb -o= 2n2d-first.out # only the first structure is processed
x3dna-dssr -i=2n2d.pdb --nmr -o=2n2d-all.out # all 10 structures are processed
x3dna-dssr -i=2n2d.pdb --nmr --json -o=2n2d-all.json # ibid., with output in JSON
Note that the NMR file is named 2n2d.pdb, and it contains 10 structures.
Interesting mixes show up when an X-ray biological assembly with multiple MODEL/ENDMDL entries is analyzed with --nmr, or an NMR entry is handled with --symmetry. Here are two such examples:
x3dna-dssr -i=2d94.pdb1 --nmr -o=temp # models 1 and 2 are handled sepatately
x3dna-dssr -i=2n2d.pdb --symm -o=temp # wrong -- does not make sense!
In summary, the --symmetry option is intended to treat symmetry-related molecules as a whole, as in a biological assembly of X-ray crystal structures. In contrast, the --nmr option aims to automate the analysis of each structure in a MODEL/ENDMDL-delineated ensemble, as in NMR structures or trajectories of MD simulations. The distinction between the two MODEL/ENDMDL usages is most clearly seen via a molecular visualization program: for example, check the figure below for 2d94.pdb1 (left) and 2n2d.pdb (right) when all frames are selected using Jmol.

JSON (JavaScript Object Notation) is a simple human-readable format that expresses data objects in name-value pairs. Over the years, it has surpassed XML to become the preferred data exchange format between applications. As a result, I’ve recently added the --json command-line option to DSSR to make its numerous derived parameters easily accessible.
The DSSR JSON output is contained in a compact one-line text string that may look cryptic to the uninitiated. Yet, with commonly available JSON parsers or libraries, it is straightforward to make sense of the DSSR JSON output. In this blogpost, I am illustrating how to parse DSSR-derived .json file via two command-line tools, jq and Underscore-CLI.
jq — lightweight and flexible command-line JSON processor
According to its website,
jq is like sed for JSON data – you can use it to slice and filter and map and transform structured data with the same ease that sed, awk, grep and friends let you play with text.
Moreover, like DSSR per se, “jq is written in portable C, and it has zero runtime dependencies.” Prebuilt binaries are available for Linux, OS X and Windows. So it is trivial to get jq up and running. The current stable version is 1.5, released on August 15, 2015.
Using the crystal structure of yeast phenylalanine tRNA (1ehz) as an example, here are some sample usages with DSSR-derived JSON output:
# Pretty print JSON
x3dna-dssr -i=1ehz.pdb --json | jq .
# Extract the top-level keys, in insertion order
x3dna-dssr -i=1ehz.pdb --json | jq keys_unsorted
# Extract parameters for nucleotides
x3dna-dssr -i=1ehz.pdb --json | jq .nts
# Extract nucleotide id and its base reference frame
x3dna-dssr -i=1ehz.pdb --json | jq '.nts[] | (.nt_id, .frame)'
Underscore-CLI — command-line utility-belt for hacking JSON and Javascript.
Underscore-CLI is built upon Node.js, and can be installed using the npm package manager. It is claimed as ‘the “swiss-army-knife” tool for processing JSON data – can be used as a simple pretty-printer, or as a full-powered Javascript command-line.’
Following the above examples illustrating jq, here are the corresponding commands for Underscore-CLI:
x3dna-dssr -i=1ehz.pdb --json | underscore print --color
x3dna-dssr -i=1ehz.pdb --json | underscore keys --color
x3dna-dssr -i=1ehz.pdb --json | underscore select .nts --color
x3dna-dssr -i=1ehz.pdb --json | underscore select .nts | underscore select '.nt_id, .frame' --color
jq or Underscore-CLI — which one to use?
As always, it depends. While jq feels more like a standard Unix utility (as sed, awk, grep etc), Underscore-CLI is better integrated into the Javascript language. For simple applications such as parsing DSSR output, either jq or Underscore-CLI is more than sufficient.
I use jq most of the time, but resort to Underscore-CLI for its “smart whitespace”. Here is an example to illustrate the difference between the two:
# z-axis of A.G1 (1ehz) base reference frame
# jq output, split in 5 lines
"z_axis": [
0.799,
0.488,
-0.352
]
# Underscore-CLI, in a more-readable one line
"z_axis": [0.799, 0.488, -0.352]

Recently, I read with great interest an article titled A context-sensitive guide to RNA & DNA base-pair & base-stack geometry by Dr. Jane Richardson, published in CCN (Computational Crystallography Newsletter, 2015, 5, 42—49). Highlighted in the article are Buckle and Propeller twist (see bottom left of the figure below), two of the angular parameters that characterize base-pair (bp) non-planarity. Particularly, I was intrigued by the “Notes on measures and figures” at the end:
Base normals were constructed in Mage (Richardson 2001) and twist torsions and buckle angles were measured from them; propeller-twists were measured as dihedral angles around an axis between N1/9 atoms.
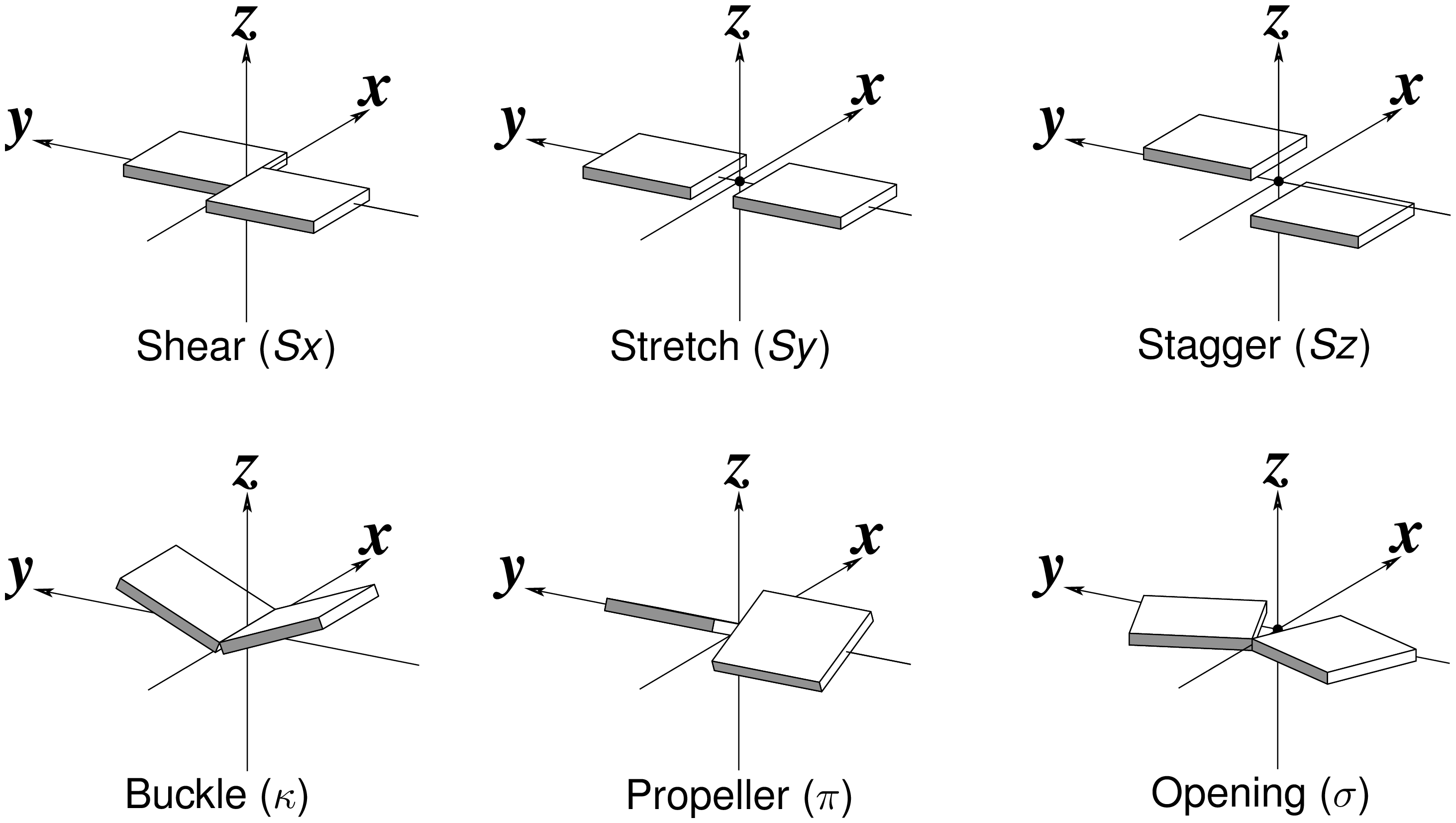
The Richardson CCN article prompted me to think more on intuitive description of bp geometry that can be easily grasped by experimentalist, especially X-ray crystallographers or cryo-EM practitioners. Without worrying about model building as with the six rigid-body parameters, it is straightforward to come up with a new set of four ‘simple’ parameters (Shear, Stretch, Buckle and Propeller) with the following characteristics:
- Each parameter can be positive or negative. For type M–N pairs (as in the canonical cases), Shear and Buckle reverse their signs when the two bases are swapped (i.e. counted as N–M instead of M–N). In all other cases, the signs of the parameters remain unchanged. See the DSSR paper for the definition of M+N vs M–N type of pairs.
- Intuitive results for non-canonical pairs, even when Opening is ~180º.
- Consistent definition between Shear/Buckle (x-axis) vs Stretch/Propeller (y-axis).
- As in 3DNA and DSSR, Buckle^2 + Propeller^2 = interBase_angle^2. Either Buckle or Propeller can render the two base planes of a pair non-parallel. Combined together, they introduce a non-zero inter-base angle. By definition, each parameter should not be larger than the overall inter-base angle.
With the cartoon-block representation introduced in DSSR, base-stacking interactions and bp deformations (especially Buckle and Propeller) are immediately obvious. Two example are illustrated in the figure below: one is the classic Dickerson B-DNA dodecamer (355d, DSSR output), and the other is the parallel double-stranded helix of poly(A) RNA (4jrd, DSSR output).
A portion of DSSR output for the B-DNA duplex 355d is shown below. Note that the first bp (at the bottom left in the figure above) has a Propeller of –17º (and a Buckle of +7º). As beautifully explained by Calladine et al. in their book Understanding DNA,
The Molecule & How It Works, Watson-Crick pairs prefer to have negative Propeller in right-handed DNA double helices to improve same-strand base-stacking interactions. The average value of Propeller in A- and B-DNA crystal structures is around –11º (see Table 3 of the Olson et al. standard base reference frame paper).
nt1 nt2 bp name Saenger LW DSSR
1 A.DC1 B.DG24 C-G WC 19-XIX cWW cW-W
[-105.9(anti) ~C2'-endo lambda=53.5] [-141.3(anti) ~C3'-endo lambda=52.7]
d(C1'-C1')=10.71 d(N1-N9)=8.96 d(C6-C8)=9.88 tor(C1'-N1-N9-C1')=-21.4
H-bonds[3]: "O2(carbonyl)-N2(amino)[2.83],N3-N1(imino)[2.90],N4(amino)-O6(carbonyl)[2.98]"
interBase-angle=19 Simple-bpParams: Shear=0.28 Stretch=-0.13 Buckle=7.3 Propeller=-17.2
bp-pars: [0.28 -0.14 0.07 6.93 -17.31 -0.61]
2 A.DG2 B.DC23 G-C WC 19-XIX cWW cW-W
[-85.4(anti) ~C2'-endo lambda=53.4] [-150.3(anti) ~C3'-endo lambda=55.4]
d(C1'-C1')=10.61 d(N1-N9)=8.92 d(C6-C8)=9.83 tor(C1'-N1-N9-C1')=-21.7
H-bonds[3]: "O6(carbonyl)-N4(amino)[2.91],N1(imino)-N3[2.88],N2(amino)-O2(carbonyl)[2.88]"
interBase-angle=17 Simple-bpParams: Shear=-0.24 Stretch=-0.18 Buckle=9.0 Propeller=-14.5
bp-pars: [-0.24 -0.18 0.49 9.34 -14.30 -2.08]
A portion of DSSR output for the parallel A-DNA duplex 4jrd is shown below. Note that the values of ‘simple’ Propeller are positive for both bps #7 and #8. In contrast, the rigid-body bp parameters have their signs flipped over when Opening is switched from –179.56º for bp#7 to +179.23º for bp#8. This sign ‘ambiguity’ around 180º Opening could be confusing. Yet, all the six bp parameters must be kept as they are for rigorous rebuilding, especially within a larger context than a bp per se. From the very beginning, 3DNA has adopted the convention of keeping angular parameters in the range of [–180º, +180º] instead of [0, 360º], allowing left-handed Z-DNA to have negative twist.
7 A.A8 B.A7 A+A -- 02-II tHH tM+M
[-175.8(anti) ~C3'-endo lambda=10.2] [-172.7(anti) ~C3'-endo lambda=12.6]
d(C1'-C1')=11.15 d(N1-N9)=8.29 d(C6-C8)=6.31 tor(C1'-N1-N9-C1')=160.1
H-bonds[4]: "OP2-N6(amino)[2.97],N7-N6(amino)[2.97],N6(amino)-OP2[2.92],N6(amino)-N7[2.91]"
interBase-angle=14 Simple-bpParams: Shear=-7.88 Stretch=0.66 Buckle=-7.8 Propeller=11.9
bp-pars: [-6.00 5.15 -0.02 0.63 14.22 -179.56]
8 A.A9 B.A8 A+A -- 02-II tHH tM+M
[-177.4(anti) ~C3'-endo lambda=12.4] [-175.8(anti) ~C3'-endo lambda=10.3]
d(C1'-C1')=11.01 d(N1-N9)=8.15 d(C6-C8)=6.18 tor(C1'-N1-N9-C1')=158.5
H-bonds[4]: "OP2-N6(amino)[2.93],N7-N6(amino)[2.88],N6(amino)-OP2[2.97],N6(amino)-N7[2.92]"
interBase-angle=15 Simple-bpParams: Shear=-7.91 Stretch=0.56 Buckle=-7.0 Propeller=13.7
bp-pars: [6.11 -5.06 -0.05 -2.26 -15.22 179.23]

From early on, the x3dna.org domain and its related sub-domains (e.g., for the forum and the web-interface to DSSR) has been served via shared hosting. By and large, this simple arrangement has worked quite well. Over the years, though, I’ve gradually realized some of its inherent limitations. One is the limited resources available to the 3DNA-related websites. Another is the accessibility issue from countries like China.
To remedy such issues, I’ve recently moved the 3DNA Forum and the web-interface to DSSR to a dedicated web server at Columbia University. Moreover, a duplicate copy of the 3DNA homepage is made available via http://home.x3dna.org hosted at Columbia. The three new websites have been verified to be accessible directly from China.
These updates on x3dna.org not only ensure global accessibility to 3DNA/DSSR, but also allow for more web services to be made available.

As of v1.3.3-2015sep03, DSSR outputs the reference frame of any base or base-pair (bp). With an explicit list of such reference frames, one can better understand how the 3DNA/DSSR bp parameters are calculated. Moreover, third-party bioinformatics tools can take advantage of the frames for further exploration of nucleic acid structures, including visualization.
Let’s use the G1–C72 bp (detailed below) in the yeast phenylalanine tRNA (1ehz) as an example:
1 A.G1 A.C72 G-C WC 19-XIX cWW cW-W
The standard base reference frame for A.G1 is:
{
rsmd: 0.008,
origin: [53.757, 41.868, 52.93],
x_axis: [-0.259, -0.25, -0.933],
y_axis: [-0.543, 0.837, -0.073],
z_axis: [0.799, 0.488, -0.352]
}
And the one for A.C72 is:
{
rsmd: 0.006,
origin: [53.779, 42.132, 52.224],
x_axis: [-0.402, -0.311, -0.861],
y_axis: [0.451, -0.886, 0.109],
z_axis: [-0.797, -0.345, 0.497]
}
The G1–C72 bp reference frame is:
{
rsmd: null,
origin: [53.768, 42, 52.577],
x_axis: [-0.331, -0.283, -0.9],
y_axis: [-0.497, 0.863, -0.089],
z_axis: [0.802, 0.418, -0.427]
}
The beauty of the DSSR JSON output is that the above information can be extracted on the fly. For example, the following commands extract the above frames:
x3dna-dssr -i=1ehz.pdb --json | jq '.ntParams[] | select(.nt_id=="A.G1") | .frame'
x3dna-dssr -i=1ehz.pdb --json | jq '.ntParams[] | select(.nt_id=="A.C72") | .frame'
x3dna-dssr -i=1ehz.pdb --json --more | jq .pairs[0].frame
Note that in JSON, the array is 0-indexed, so the first bp (G1–C72) has an index of 0. In addition to jq, I also used underscore to pretty-print the frames.