In the field of nucleic acid structures, especially in the ‘RNA world’, we often hear named base pairs (bp). Among those, the Watson-Crick (WC) A–U and G–C bps (see figure below) are by far the most common.
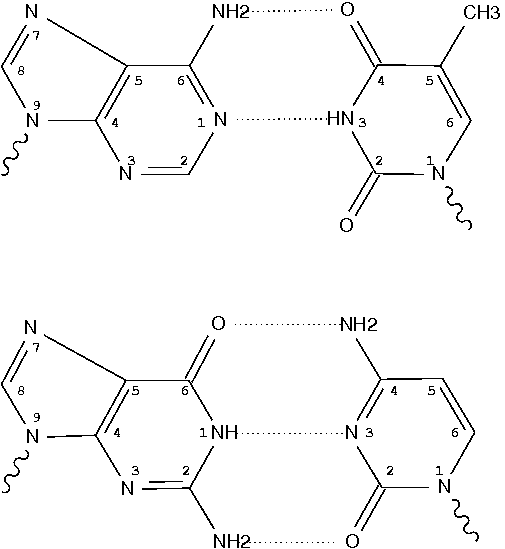
Reversed WC (rWC) base pairs
Closely related to the WC bps are the so-called reversed WC (rWC) bps, where the relative glycosidic bond are reversed; instead of being on the same side of the bases as in WC bps shown above, they are now on opposite sides in rWC bps as shown below. According to the Leontis-Westhof (LW) bp classification scheme, the rWC bps belong to trans WC/WC. Following Saenger’s numbering, the rWC A+U bp corresponds to XXI, and the rWC G+C bp XXII.
In the figures below, the name of each type of bp and its LW & Saenger designations (separated by ‘;’) are noted under the corresponding image. All images are generated with 3DNA; for easy comparison, each bp is oriented in the reference frame of the leading base.
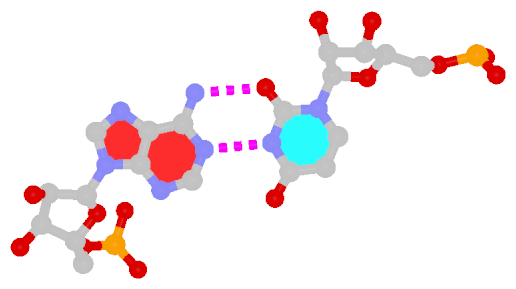 |
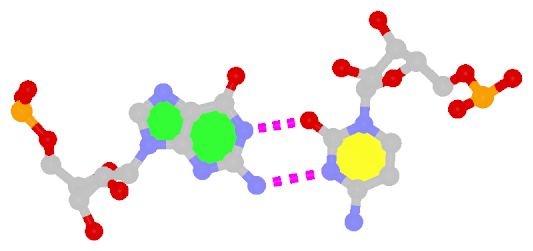 |
| Reversed WC A+U pair | Reversed WC G+C pair |
| trans WC/WC; XXI | trans WC/WC; XXII |
Hoogsteen and reversed Hoogsteen base pairs
The next most famous one is the Hoogsteen A+U bp, which also has a reverse variant, i.e., the rHoogsteen A–U bp (see figure below). Now the major groove edge of A, termed the Hoogsteen edge by LW, is used for pairing with U.
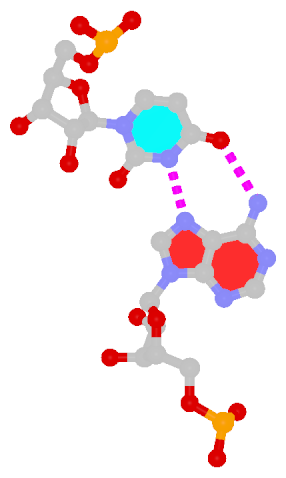 |
 |
| Hoogsteen A+U pair | Reversed Hoogsteen A–U pair |
| cis Hoogsteen/WC; XXIII | trans Hoogsteen/WC; XXIV |
The G–U Wobble base pair
First proposed by Crick in 1966 to account for the degeneracy in codon–anticodon pairing, the Wobble bp is an essential component (in addition to the WC bps) in forming double helical RNA secondary structures.
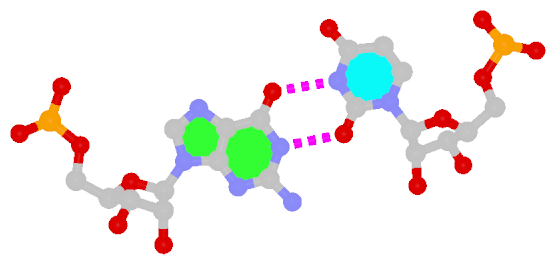 |
| Wobble G–U pair |
| cis WC/WC; XXVIII |
The sheared G–A base pair
Sheared G–A is a commonly found non-WC bp in both DNA and RNA structures. Noticeably, tandem sheared G–A bps introduce distinct stacking geometry. Here G uses its minor groove edge, termed the sugar edge by LW, to pair with the Hoogsteen edge of A.
 |
| Sheared G–A pair |
| trans Suger/Hoogsteen; XI |
Dinucleotide platforms
Dinucleotide platforms are formed via side-by-side pairing of adjacent bases; the most common of which are GpU and ApA. Here the sugar (minor-groove) edge of the 5′ base interacts with the Hoogsteen (major-groove) edge of the 3′ base. Since there is only one base-base H-bond in dinucleotide platforms, no Saenger classification is available. In 3DNA output, the GpU dinucleotide platform is designated as G+U, and ApA as A+A.
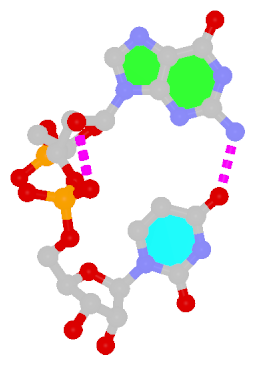 |
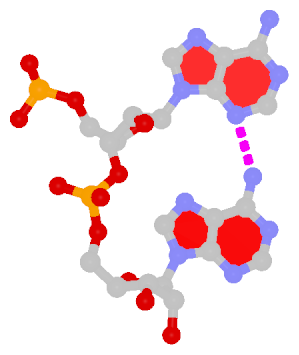 |
| GpU dinucleotide platform | ApA dinucleotide platform |
| cis Sugar/Hoogsteen; n/a | cis Sugar/Hoogsteen; n/a |
Other named base pairs
There exist other named bps in RNA literature, e.g., G⋅A imino, A⋅C reverse Hoogsteen, G⋅U reverse Wobble etc. In the my experience, they are (much) less commonly used than the ones illustrated above.
